|
|
| (11 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Team:Newcastle/mainbanner}} | | {{Team:Newcastle/mainbanner}} |
| - | '''Tuesday'''
| |
| - | ===Aims===
| |
| - | The aim of today's Lab practice was to extract the plasmids for GFP and RFP, digest the 2 plasmids and extract the 2 inserts and 1 of the backbones (vector)from a gel. From this a ligation was set up.
| |
| | | | |
| - | ===Equipement List=== | + | ==Aims== |
| | + | The aim of today's Lab practice was to extract the plasmids for GFP and RFP, digest the 2 plasmids and extract the 2 inserts and 1 of the backbones (vector)from an agarose gel. From this a ligation was set up. |
| | | | |
| - | For today's Lab pratice we required:
| + | ==Materials and Protocol== |
| | | | |
| - | * Broth cultures from yesterday
| + | ===QIagen Miniprep Kit Using a Microcentrifuge for Plasmid Extraction=== |
| - | * Solutions for Minipreps( plasmid and gel extraction,digest and ligation)
| + | Please refer to: [[Team:Newcastle/Qiagen Minipreps|Qiagen Minipreps]] for materials required and protocol. |
| - | * Buffers
| + | |
| - | * Agarose gel
| + | |
| - | * Microwave
| + | |
| - | * Restriction enzymes
| + | |
| - | * Gloves(for handling the gel)
| + | |
| - | * Scalpel
| + | |
| - | * Scales
| + | |
| - | * Transilluminator
| + | |
| - | * Nanodrop
| + | |
| - | | + | |
| - | | + | |
| - | ===Qiagen miniprep: Plasmid extraction(Summary)===
| + | |
| - | The 5ml culture set up yesterday was pelleted down using the microcentrifuge, the pellet was resuspended in buffer P1.
| + | |
| - | Lysis buffer P2 was used to release the DNA from the cell (this buffer contains RNAse to help reduce RNA contamination). PE buffer containing ethanol was used as a wash. We lysed the cells for 1 minute, gently inverting the tube. We neutalised the lysis with N3 buffer and centrifuged for 10minutes, only the '''plasmid DNA was left in suspension'''. The DNA was eluted at low salt concentration through a column membrane.
| + | |
| - | | + | |
| - | ====Miniprep Kit Using a Microcentrifuge:Step by step====
| + | |
| - | The Miniprep Kit is used to extract plasmid, which is the first thing that we did.
| + | |
| - | # Resuspend pelleted bacterial cells in 250 μl Buffer P1 and transfer to a microcentrifuge tube.
| + | |
| - | # Add 250 μl Buffer P2 and mix throughly by inverting the tube 4-6 times.
| + | |
| - | # Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
| + | |
| - | # Centrifuge for 10 min at 13,000 rpm in a table-top microcentrifuge.
| + | |
| - | # Apply the supernatant to the QIAprep spin column by decanting or pipetting.
| + | |
| - | # Centrifuge for 30-60s. Discard the flow-through.
| + | |
| - | # Recommended: Wash the QIAprep spin column by adding 0.5 ml Buffer PB and centrifuging for 30-60s. Discard the flow-through.
| + | |
| - | # Wash QIAprep spin column by adding 0.75 ml Buffer PE and centrifuging for 30-60s.
| + | |
| - | # Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer.
| + | |
| - | # To elute DNA, place the QIAprep column in a clean 1.5 ml microcentrifuge tube. Add 50 μl Buffer EB or water to the center of each QIAprep spin column, let stand for 1 min, and centrifuge for 1 min.
| + | |
| | | | |
| | ===Digest=== | | ===Digest=== |
| - | | + | Please refer to: [[Team:Newcastle/Restriction digests| Restriction digests]] for materials required and protocol. |
| - | Cut with restriction enzyme ''Eco''R1 and ''Pst''1 1 μl of each, the prefix and suffix of the BBa_04450 biobrick.
| + | |
| - | | + | |
| - | 10*buffer ... so 3 μl in 30μ1 therefore we can have 25μ1 of DNA
| + | |
| - | Also no more than 10% Glycerol in the enzyme solution or the reaction would be inhibited.
| + | |
| - | | + | |
| - | # 20 μl solution should be made in total: 15 μl plasmid (with GFP/RFP), 1 μl EcoRI, 1 μl PstI, 2 μl Buffer (10x), 1μl water. '''NOTE:''' Digestive enzymes should never exceed 10% of the total volume.
| + | |
| - | # Add the reagents in this order: plasmid, water, buffer, enzymes.
| + | |
| - | # Centrifuge for a few seconds to make sure that the mixture is at the bottom.
| + | |
| - | # Heat block for 2 hours at 37 degrees C.
| + | |
| | | | |
| | ===Gel extraction=== | | ===Gel extraction=== |
| - | | + | Please refer to: [[Team:Newcastle/Gel extraction| Gel extraction]] for materials required and protocol. |
| - | Gel electrophoresis can be used to separate DNA, RNA, or protein molecules by molecular weight using an electric field applied to a gel matrix
| + | |
| - | | + | |
| | | | |
| - | ====Agarose gel====
| + | ===Agarose Gel Electrophoresis=== |
| - | | + | Please refer to: [[Team:Newcastle/Gel electrophoresis| Gel electrophoresis]] for materials required and protocol. |
| - | Agarose gel is used for DNA seperation. 1% agarose(1g of agarose in 100 ml of TAE buffer) is used because it is suitable for most kilobase pairs of DNA. We used 60ml for our gel. We used SafeView dye (3 μl) to bind the DNA so that the DNA is visible. We used TAE buffer and boiled the mixture in a microwave to melt it. We then let it cool and poured it into the tank to set( Wait for 30 min to allow the gel to harden).
| + | |
| - | | + | |
| - | ====Electrophoresis====
| + | |
| - | | + | |
| - | Transfer harden gel into the gel tank and add TAE buffer until the gel is completely submerge
| + | |
| - | Depending on the nature of the sample, 3μl of GeneRuler™ 1kb Plus DNA Ladder was used for analysing DNA
| + | |
| - | Loading buffer was then added together with the sample before loading onto the gel matrix
| + | |
| - | Run gel at 90V until separation is achieved and visualize using the gelDoc
| + | |
| | | | |
| - | In lane one we loaded the molecular marker 5μl. In lane three we loaded the digested RFP plasmid. In lane five we loaded the digested GFP plasmid. In lane eight we loaded 5μl RFP plasmid and 1μl sample buffer. Lanes two, four, six and seven are empty.
| |
| | {| | | {| |
| | |- | | |- |
| - | |[[Image:Newcastle_lab_1.jpeg|thumb]] | + | |[[Image:Newcastle_lab_1.jpeg|thumb|Dr Wendy demostrating how to use gel electrophoresis machine]] |
| - | |[[Image:Newcastle_lab_2.jpeg|thumb]] | + | |[[Image:Newcastle_lab_2.jpeg|thumb|Gel tank]] |
| - | |[[Image:Newcastle_lab_3.jpeg|thumb]] | + | |[[Image:Newcastle_lab_3.jpeg|thumb|Deena pouring hot molten gel]] |
| | |- | | |- |
| | |} | | |} |
| Line 79: |
Line 30: |
| | {| | | {| |
| | |- | | |- |
| - | |[[Image:Newcastle_lab_4.jpeg|thumb]] | + | |[[Image:Newcastle_lab_4.jpeg|thumb|Putting the gel on the geldoc]] |
| - | |[[Image:Newcastle_lab_5.jpeg|thumb]] | + | |[[Image:Newcastle_lab_5.jpeg|thumb|Preparing to cut the gel]] |
| - | |[[Image:Newcastle_lab_6.jpeg|thumb]] | + | |[[Image:Newcastle_lab_6.jpeg|thumb|Gel illuminated by the UV light which is emitted by the geldoc]] |
| | |- | | |- |
| | |} | | |} |
| | | | |
| - | We cut the inserts which were about 900bp and we use the backbone from the RFP plasmid. | + | We cut the inserts which were about 900bp and we use the backbone from the plasmid consisting ''rfp''. |
| | | | |
| | ====QIAquick Gel extraction kit==== | | ====QIAquick Gel extraction kit==== |
| Line 91: |
Line 42: |
| | {| | | {| |
| | |- | | |- |
| - | |[[Image:Newcastle_lab_7.jpeg|thumb]] | + | |[[Image:Newcastle_lab_7.jpeg|thumb|Phil cutting the gel]] |
| - | |[[Image:Newcastle_lab_8.jpeg|thumb]] | + | |[[Image:Newcastle_lab_8.jpeg|thumb|Transferring the cut gel part into a 2 ml microfuge tube]] |
| | |- | | |- |
| | |} | | |} |
| | | | |
| - | The DNA fragments are cut from the agarose gel with a scalpel. Three parts of QG buffer is added to one part of gel and the mixtures are dissolved at the temperature of 50°C. The tubes are vortexed every 2-3 minutes to help dissolve the gel. 1 part of isopropanol is added to the sample and mix. The microcubes were spinned in the microfuge for 1 minute.
| + | ==Set up ligation== |
| - | | + | |
| - | ====QIAquick gel extraction microcentrifuge protocol====
| + | |
| - | # Excise the DNA fragment from the agarose gel with a clean, sharp scalpel
| + | |
| - | # Weight the gel slice and add 3 volumes of buffer QG to 1 volume of gel (100 mg ~ 100 µl)
| + | |
| - | # Incubate at 50°C and invert the tube gently at regular intrerval until the gel has completely dissolved
| + | |
| - | # After the gel has dissovled completely, check that the color of the mixture is yellow
| + | |
| - | # Add 1 gel volume of isopropanol to the sample and mix
| + | |
| - | # Place a QIAquick spin column in a 2 ml collection tube
| + | |
| - | # To bind DNA, apple the sample to the QIAquick column and centrifuge for 1 min
| + | |
| - | # Discard the flow through and place the QIAquick column back into the same tube (Maximum volumn is 750ul)
| + | |
| - | # Add 0.5 ml of Buffer QG to QIAquick column and centrifuge for 1 min and discard the flow through and place the QIAquick column back into the same tube
| + | |
| - | # To wash, add 0.75 ml of Buffer PE to QIAquick column and centrifuge for 1 min and place the QIAquick column back into the same tube
| + | |
| - | # Centrifuge the column for a further 1 min
| + | |
| - | # Transfer the column into a clean 1.5 ml micriocentrifuge tube
| + | |
| - | # To elute DNA, add 30 µl of Buffer EB to the center of the QIAquick membrane and allow to stand for 1 min
| + | |
| - | # Centrifuge the column for 1 min and transfer the eluate to a clean 1.5 ml micriocentrifuge tube
| + | |
| - | | + | |
| - | ====Weighing====
| + | |
| - | | + | |
| - | All gel extractions weighed 0.3g.
| + | |
| - | | + | |
| - | ===Set up ligation===
| + | |
| | | | |
| | {|border=1 | | {|border=1 |
| Line 162: |
Line 91: |
| | |10.0 | | |10.0 |
| | |} | | |} |
| - | ===Transformation protocol===
| + | ==Transformation protocol== |
| - | # Thaw a 200µl aliquot of the desired strain of ''E. coli'' and add the transforming DNA (10 ng of vector DNA in 10 µl). Incubate for 45 min at 42°C.
| + | Please refer to: [[Team:Newcastle/Transformation of E. coli|Transformation of ''E. coli'']] for materials required and protocol. |
| - | # Heat-shock the cells for 120 secs, and place on ice again for 3-4 min.
| + | |
| - | # Add 1ml of LB broth and incubate the cells at 37°C for 1-15 hr in a water bath with gentle shaking.
| + | |
| - | # Plate out 100-200 µl/plate on LB (agar at 1.5%), containing the appropriate selection markers.
| + | |
| - | # Incubate plates overnight at 37°C.
| + | |
| - | | + | |
| - | ===Nanodrop Protocol===
| + | |
| - | | + | |
| - | Nanodrop can be used to measure the DNA, RNA and protein
| + | |
| | | | |
| - | # Select Nanodrop program from the desktop
| + | ==Nanodrop Protocol== |
| - | # To clean Nanodrop, add a drop of water on the spectrometer and press blank
| + | Please refer to: [[TeamNewcastleNanoDrop Spectrophotometer| Nanodrop Spectrophotometer]] for materials required and protocol. |
| - | # After cleaning, wipe the water off
| + | |
| - | # To equalizen the equipment, add 3 μl of the buffer used in the sample and press blank
| + | |
| - | # Wipe the buffer off
| + | |
| - | # To measure sample, add 3 μl of the sample and press measure
| + | |
| - | # If dealing with multiple samples, clean the equipment with water at regular intervals
| + | |
| - | # After measurement, clean the equipment with a drop of water on the spectrometer and press blank
| + | |
| | | | |
| | | | |
| | {| | | {| |
| | |- | | |- |
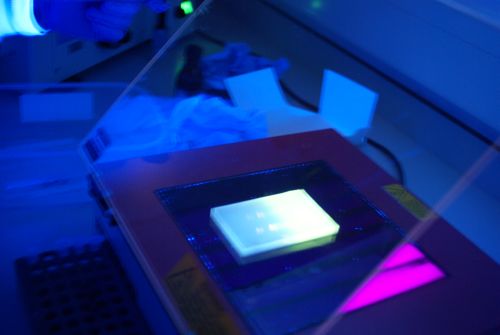
| - | |[[Image:Newcastle_nanodrop_1.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_1.jpeg|thumb|A typical curve to be achieved from a nanodrop spectrophotmeter]] |
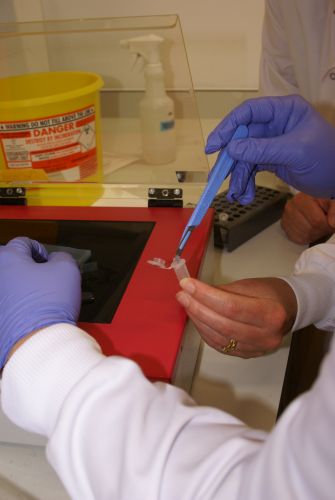
| - | |[[Image:Newcastle_nanodrop_2.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_2.jpeg|thumb|Phil analyzing a sample on the nanodrop machine]] |
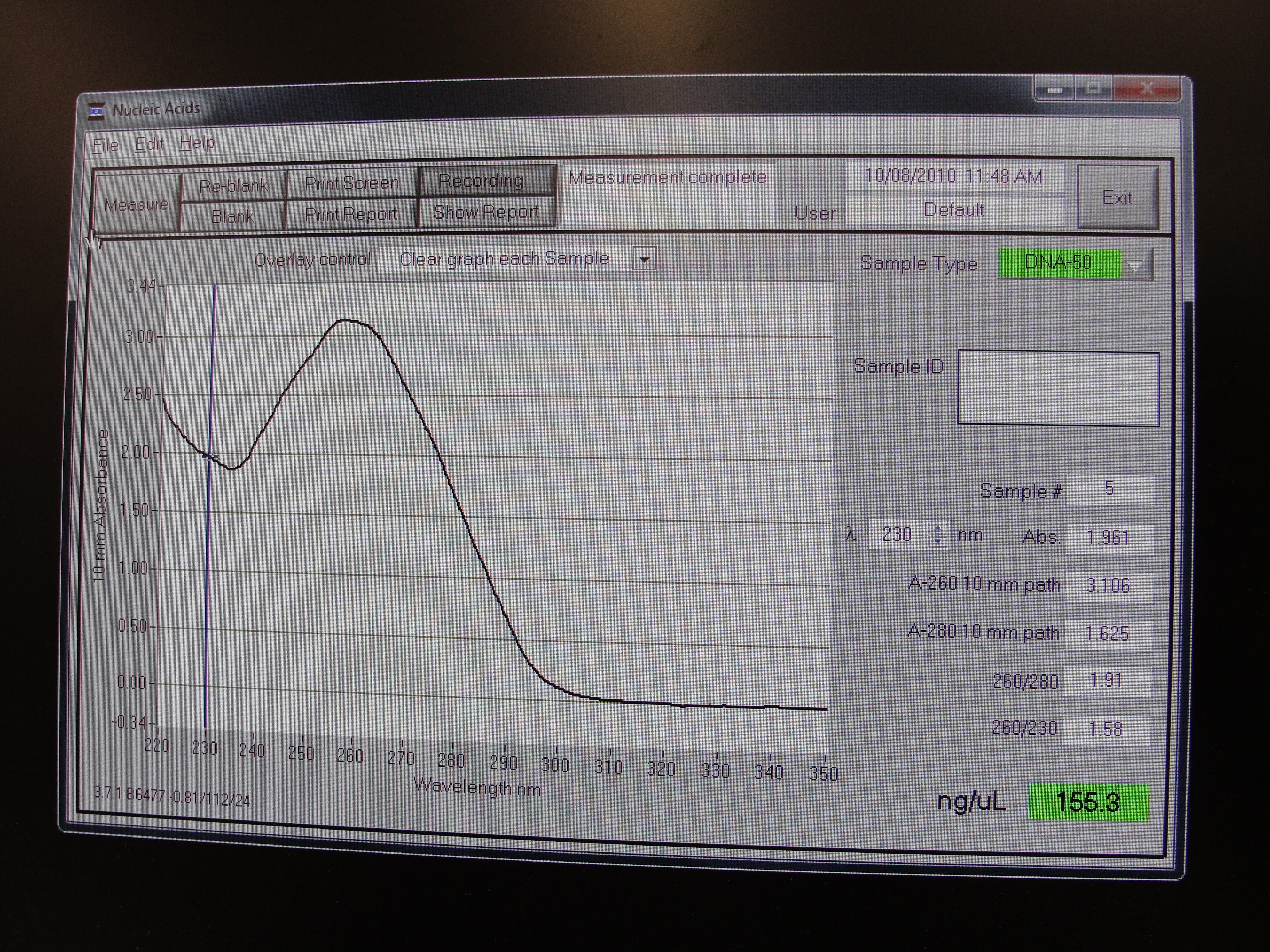
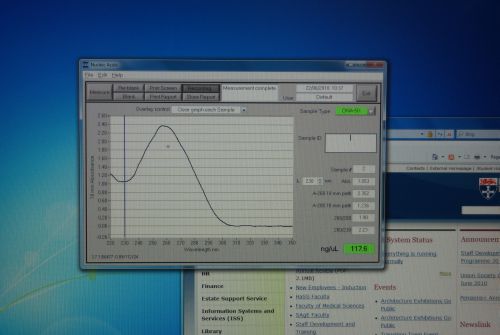
| - | |[[Image:Newcastle_nanodrop_3.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_3.jpeg|thumb|Analyzed data on the screen]] |
| | |- | | |- |
| | |} | | |} |



 "
"