Team:Newcastle/13 August 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) (→Result) |
Shethharsh08 (Talk | contribs) (→Result) |
||
| Line 58: | Line 58: | ||
The expect band size is supposed to be 3195 bp after Gibson procedure ligation. In the Figure 1, we can see that in Lane 2 and 5 we got a band of approximately 3200 bp but we found that it is because of the initial insert (''rfp'' gene) which was present in the plasmid pSB1C3 i.e. these 2 sequences are of the template DNA which were used during PCR reaction few days ago. The other 4 bands are of approximately 2100 bp in size. | The expect band size is supposed to be 3195 bp after Gibson procedure ligation. In the Figure 1, we can see that in Lane 2 and 5 we got a band of approximately 3200 bp but we found that it is because of the initial insert (''rfp'' gene) which was present in the plasmid pSB1C3 i.e. these 2 sequences are of the template DNA which were used during PCR reaction few days ago. The other 4 bands are of approximately 2100 bp in size. | ||
| - | == | + | ==Conclusion== |
Overall the GIbson protocol has failed. We did not receive bands of correct size thus showing that the ''rfp'' fragments and the plasmid pSB1C3 did not ligate at all. The 2 bands of approximately 3200 bp size are of the plasmid pSB1C3 containing ''rfp'' gene insert and thus these fragments are of template DNA for the PCR reaction which had taken place few days ago. The explanation for the failure of the Gibson protocol is as following: | Overall the GIbson protocol has failed. We did not receive bands of correct size thus showing that the ''rfp'' fragments and the plasmid pSB1C3 did not ligate at all. The 2 bands of approximately 3200 bp size are of the plasmid pSB1C3 containing ''rfp'' gene insert and thus these fragments are of template DNA for the PCR reaction which had taken place few days ago. The explanation for the failure of the Gibson protocol is as following: | ||
# Because of the presence of the BioBrick prefix and suffix sequences at the ends of the fragments, the fragments circularized and ligated with themselves. This is because both prefix and suffix contains Not1 restriction site and Xba1 and Spe1 restriction site which have similar sequences and thus during ligation step of the Gibson reaction, the fragments containing prefix and suffix religate with themselves and thus the fragments have not been able to ligate with each other as expected earlier. | # Because of the presence of the BioBrick prefix and suffix sequences at the ends of the fragments, the fragments circularized and ligated with themselves. This is because both prefix and suffix contains Not1 restriction site and Xba1 and Spe1 restriction site which have similar sequences and thus during ligation step of the Gibson reaction, the fragments containing prefix and suffix religate with themselves and thus the fragments have not been able to ligate with each other as expected earlier. | ||
Revision as of 14:14, 18 August 2010

| |||||||||||||
| |||||||||||||
Contents |
rocF BioBrick miniprep
Aims
The aim of this experiment is to extract pSB1C3 containing the rocF BioBrick from E. coli DH5α cells with the help of Qiagen miniprep kit and confirming the extraction with the help of a nanodrop experiment.
Materials and Protocol
Please refer to: Minipreps for Qiagen miniprep protocol, Nanodrop Spectrophotometer for nanodrop protocol.
Result
| pSB1C3 with rocF BioBrick (no. 1) | pSB1C3 with rocF BioBrick (no. 2) | pSB1C3 with rocF BioBrick (no. 3) | pSB1C3 with rocF BioBrick (no. 4) | pSB1C3 with rocF BioBrick (no. 5) | pSB1C3 with rocF BioBrick (no. 6) |
|---|---|---|---|---|---|
| 90.3 µl/ml | 64.4 µl/ml | 286.9 µl/ml | 87.1 µl/ml | 780.5 µl/ml | 59.1 µl/ml |
Table 1: Nanodrop spectrophotometer experiment result. Table represents the amount of plasmid present in µl/ml quantity.
Discussion
The tube containing plasmid pSB1C3 with rocF BioBrick (no.3) has the highest concentration with a high purity value.
Conclusion
Overall the plasmid miniprep was successful and we would be using Tube no. 3 containing plasmid pSB1C3 with rocF BioBrick.
rocF BioBrick gel electrophoresis
Aims
The aim of this experiment is to run gel electrophoresis for the extracted plasmid pSB1C3 containing rocF BioBrick.
Materials and Protocol
Please refer to: Gel electrophoresis for gel electrophoresis protocol.
Result
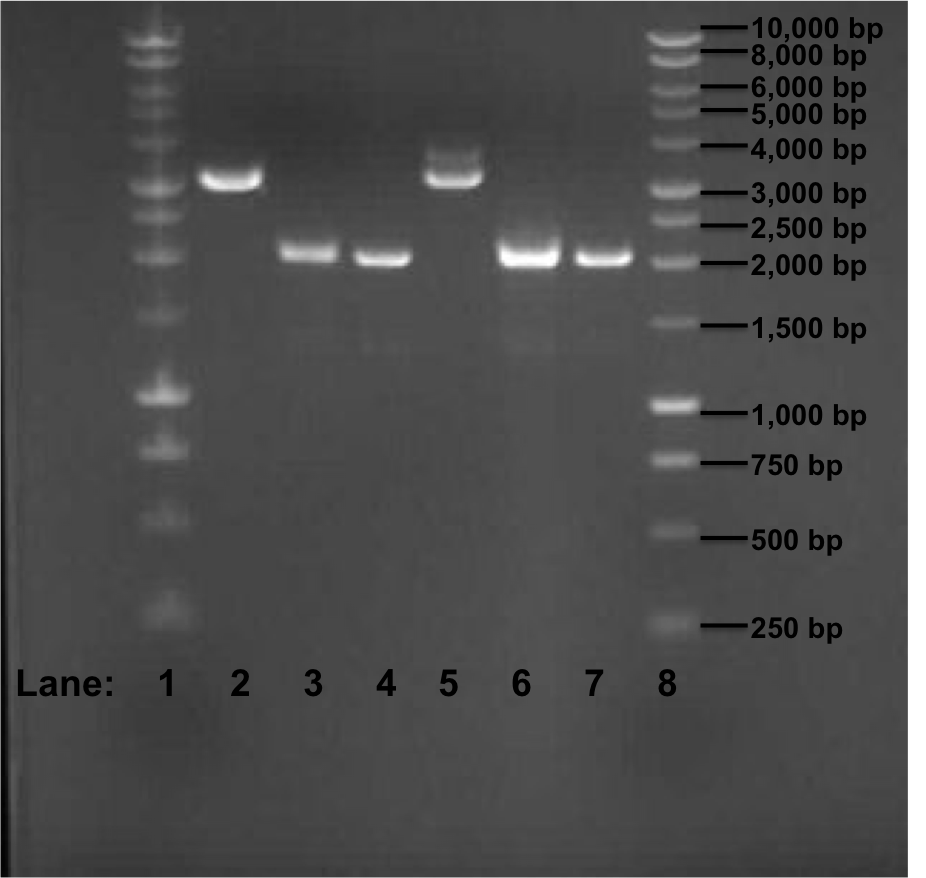
Figure 1: Gel electrophoresis of the miniprep products required for the confirmation of Gibson procedure.
- Lane 1: 1 kb DNA ladder
- Lane 2: pSB1C3 with rocF BioBrick (no. 1)
- Lane 3: pSB1C3 with rocF BioBrick (no. 2)
- Lane 4: pSB1C3 with rocF BioBrick (no. 3)
- Lane 5: pSB1C3 with rocF BioBrick (no. 4)
- Lane 6: pSB1C3 with rocF BioBrick (no. 5)
- Lane 7: pSB1C3 with rocF BioBrick (no. 6)
- Lane 8: 1 kb DNA ladder
Discussion
The expect band size is supposed to be 3195 bp after Gibson procedure ligation. In the Figure 1, we can see that in Lane 2 and 5 we got a band of approximately 3200 bp but we found that it is because of the initial insert (rfp gene) which was present in the plasmid pSB1C3 i.e. these 2 sequences are of the template DNA which were used during PCR reaction few days ago. The other 4 bands are of approximately 2100 bp in size.
Conclusion
Overall the GIbson protocol has failed. We did not receive bands of correct size thus showing that the rfp fragments and the plasmid pSB1C3 did not ligate at all. The 2 bands of approximately 3200 bp size are of the plasmid pSB1C3 containing rfp gene insert and thus these fragments are of template DNA for the PCR reaction which had taken place few days ago. The explanation for the failure of the Gibson protocol is as following:
- Because of the presence of the BioBrick prefix and suffix sequences at the ends of the fragments, the fragments circularized and ligated with themselves. This is because both prefix and suffix contains Not1 restriction site and Xba1 and Spe1 restriction site which have similar sequences and thus during ligation step of the Gibson reaction, the fragments containing prefix and suffix religate with themselves and thus the fragments have not been able to ligate with each other as expected earlier.
Subtilin Immunity BioBrick
Aims
We plan to select three spaIFEG gene (part 3) PCR tubes, using Tms 51°C, 56°C and 61°C (we had confirmed it successful from yesterday's gel electrophoresis - please refer to the gel electrophoresis from yesterday). Gel extraction will then be performed, along with Plasmid Vector (part 1), Promoter & RBS (part 2) and Double terminator (part 4). Finally, NanoDrop will be performed on each of these four parts.
Materials and protocol
Please refer to the gel electrophoresis, gel extraction and NanoDrop protocols. Instead of using the centrifuge to bind the DNA from our sample to the QIAquick column, a vacuum manifold was used instead. It is the first time it had been used. The advantage of this was that the extraction could be carried out much faster as we had 7 samples. (photos)
Results
The results from the Nanodrop Spectrophotometer of the four parts are as follows:
- Vector - 21.3 ng/μl
- Promoter - 28.8 ng/μl
- Coding sequence - 27.0 ng/μl
- Terminator - 27.5 ng/μl
Discussion
We chose the three spaIFEG gene (part 3) PCR tubes, using Tms 51°C, 56°C and 61°C because they were the mid-ranged Tms. Gel extraction was then performed, by firstly cutting the gels under U.V light, and then apply the protocol for completion.
Conclusion
The results from the NanoDrop reading suggests that our results were positive, which indicates that we have successfully extracted all the parts required for the Subtilin Immunity BioBrick. Gibson cloning method will be used next week.
Go back to our main Lab book page
 
|
 "
"