Team:Newcastle/11 August 2010
From 2010.igem.org
(Difference between revisions)
(→Gibson cloning of the rocF BioBrick) |
(→Discussion) |
||
| (5 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | =Gel electrophoresis for the subtilin immunity fragments= | |
| - | + | ||
| - | = | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Aims== | ==Aims== | ||
| - | + | The aim of the experiment is to check whether we have the correct fragment sizes on the four subtilin fragments that are we have amplified [[Team:Newcastle/10 August 2010#Subtilin_Immunity_BioBrick| yesterday]]. | |
| - | + | ||
| - | + | ||
==Materials and protocol== | ==Materials and protocol== | ||
| - | Please refer to the [[Team:Newcastle/Gel_electrophoresis| gel electrophoresis]] | + | Please refer to the [[Team:Newcastle/Gel_electrophoresis| gel electrophoresis]] protocol. |
==Results== | ==Results== | ||
| Line 47: | Line 29: | ||
==Discussion== | ==Discussion== | ||
| - | Plasmid Vector (lane 2), Promoter & RBS (lane 3) and Double terminator (lane 5) | + | The correct band size were observed for for the Plasmid Vector (lane 2), Promoter & RBS (lane 3) and Double terminator (lane 5). However no band was observed for the ''spaIFEG''(lane 4). |
| - | ''spaIFEG'' | + | |
| + | ==Conclusion== | ||
| + | No band was observed for the ''spaIFEG''. This could be due to the wrong melting temperature. | ||
'''Go back to our main [[Team:Newcastle/notebook| Lab book]] page''' | '''Go back to our main [[Team:Newcastle/notebook| Lab book]] page''' | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 00:11, 26 October 2010

| |||||||||||||
| |||||||||||||
Contents |
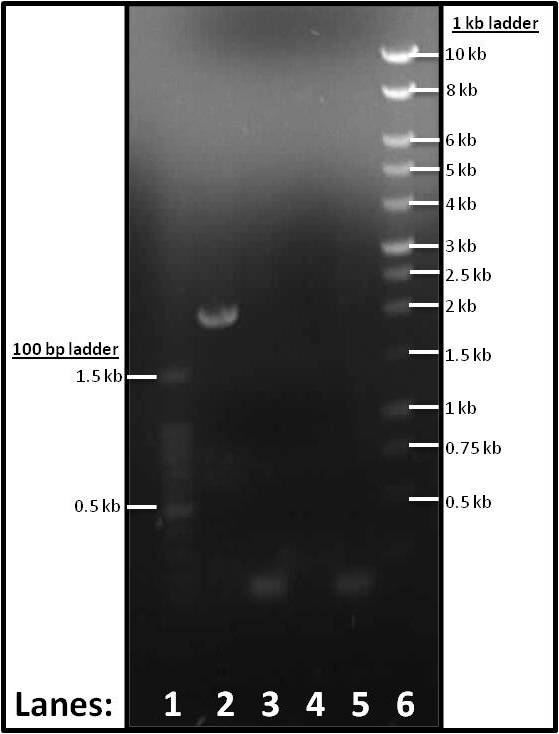
Gel electrophoresis for the subtilin immunity fragments
Aims
The aim of the experiment is to check whether we have the correct fragment sizes on the four subtilin fragments that are we have amplified yesterday.
Materials and protocol
Please refer to the gel electrophoresis protocol.
Results
Figure 1: Gel electrophoresis of the PCR products of the parts required for the subtilin immunity BioBrick.
- Lane 1: 1 kb DNA ladder
- Lane 2: Plasmid Vector (pSB1C3)
- Lane 3: Promoter and RBS (pVeg-SpoVG)
- Lane 4: spaIFEG Gene Cluster
- Lane 5: Double terminator
- Lane 6: 100 bp DNA ladder
Discussion
The correct band size were observed for for the Plasmid Vector (lane 2), Promoter & RBS (lane 3) and Double terminator (lane 5). However no band was observed for the spaIFEG(lane 4).
Conclusion
No band was observed for the spaIFEG. This could be due to the wrong melting temperature.
Go back to our main Lab book page
 
|
 "
"