Team:Nevada/ccdB
From 2010.igem.org
ccdB
 ccdB minimal gene Team:Nevada/registry submissions
ccdB minimal gene Team:Nevada/registry submissions

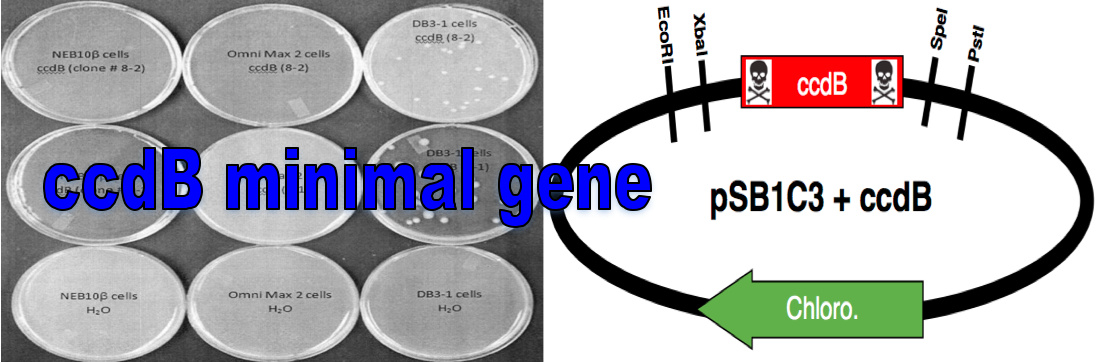
The ccdB gene is a known topoisomerase II poison and will kill most commercially available E. coli cell lines. The ccdB gene can be used as a selectable marker by placing it in the multicloning region of a plasmid and propagating it in ccdB resistant cell lines (e.g. DB3.1 from New England Biolabs). During cloning, the ccdB gene can be switched out for your gene of interest and transformed into a cell line that is susceptible to the toxic effects of ccdB (e.g NEBβ cells from New England Biolabs). Colonies that grow on plates should contain the plasmid and your gene of interest. If the ccdB gene is still present in the plasmid, the plasmid will kill any colonies where it is still present. This is an improvement to the current ccdB cell death gene part (BBa_P1010) as it does not contain the inactive ccdA gene or an unknown stuffer region.
Our Method: ccdB was amplified using Polymerase Chain Reaction (PCR) with primers designed to contain the specific BioBrick prefix and suffix. The amplified fragment was then TOPO-cloned into a PCR-Blunt II vector (Invitrogen). The vector was then cut with EcoRI and PstI restriction enzymes and the ccdB fragment was transformed into the iGEM compatible vector pSB1C3.
Our Results:
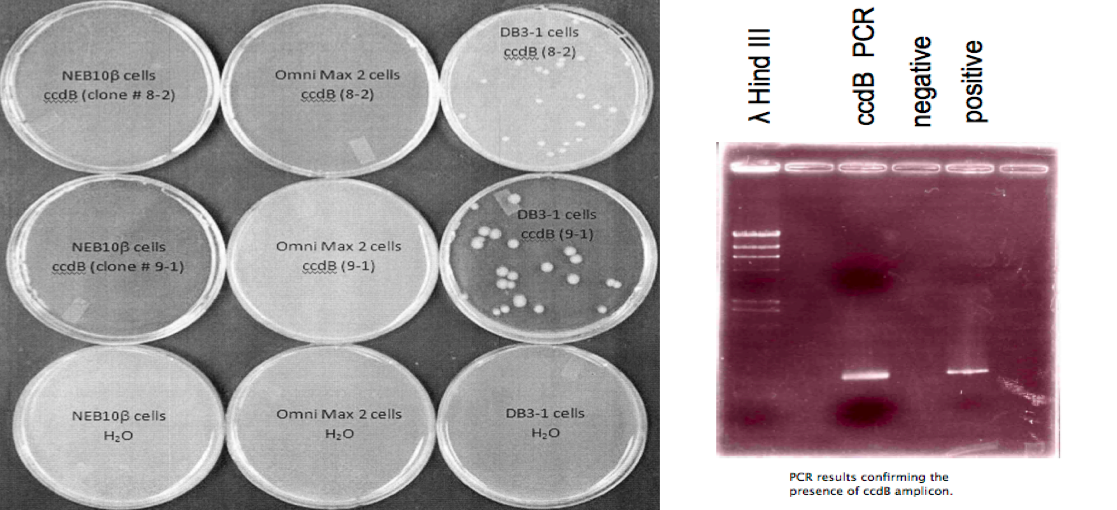
- ccdB in the PCR-Blunt II vector were transformed in three different cell lines: NEB10β cells (New England Biolabs), Omni Max 2 cells (Invitrogen), and DB3.1 cells (New England Biolabs). NEB10β and Omni Max 2 cell lines are not resistant to the toxic properties of ccdB. No colonies were present in those two cell lines but were present in DB3.1 cell lines (Right).
Vectors Used:


| 
| 
| 
| 
|
|---|
 "
"