Team:Michigan/Project
From 2010.igem.org
Overall project
Our project has several different tracks that we will be working on in the wet-lab simultaneously. These tracks include quorum sensing, pili hyperexpression, and surface display. We made the decision to work in several tracks in order to maximize the efficiency of our lab teams. Our goal is to be able to combine all of these parts in the end and form an organism that will be able to effectively flocculate microalgae.
Pili Hyperexpression
Type 1 pili (also known as fimbriae) are proteinaceous adhesins that are found on the surface of E. coli. One cell can contain up to 100 pili, which can form up to 2 um long. The pili help E. coli. form biofilms.
The pili are controlled by the fim operon. This operon consists of several genes. The pili themselves are composed of several thousand subunits of FimA. The tip of each pili consists of the genes FimF, FimG, and FimH. FimH is an adhesin and is linked to FimA through FimF and FimG. Inside the cell, FimC carries proteins to the structural platform, FimD, which then assembles the pilus rod. You can see this visually in Fig. 1 [1]. This whole process is regulated by the recombinases FimB and FimE. These genes control an invertible DNA sequence, which, when in the "on" position, promotes the production of pili. You can learn more about the regulatory system in the modeling section.
By overproducing the pili, we hope to increase flocculation. We plan to accomplish this goal by putting the FimB gene on a plasmid. Because of the way the pili regulation system works, this should promote piliation in the cell, and therefore, increase flocculation. You can find the pili team's lab notebook here.
Quorum Sensing
Our cells will use quorum sensing to determine when the flocculation will start. The cells will produce an inducer either AHL or AI-2. You can find their lab notebook here.
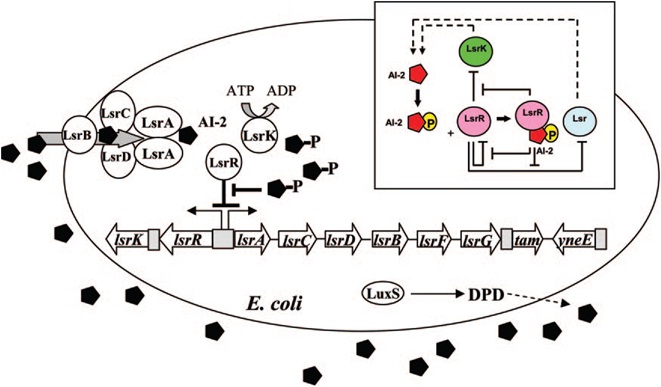
E. coli cells detect their population density by a method known as "quorum sensing." All cells produce a molecule known as Auto-Inducer 2 (AI-2) via the gene LuxS. AI-2 is actively imported via a complex of Lsr membrane proteins, after which it is phosphorylated and binds to the repressor LsrR. This binding relieves the Lsr operon from repression, increasing levels of both the AI-2 import and phosphorylation and of LsrR. The net effect is that cells respond to a threshold concentration of AI-2 (a threshold population density) by altering expression of many genes. E. coli, for example, tends to increase biofilm production in response to AI-2.
Certain species of microalgae useful for harvesting as biofuel, specifically Chlorella vulagris, have been characterized as releasing compounds that mimic the action of AI-2 or AHL, activating the quorum sensing circuits of many bacteria, including Vibrio harveyi, which responds to the same compound as E. coli, AI-2.
The goals of the quorum sensing team are two-fold: first to characterize the response of E. coli to the C. vulgaris AI-2 mimic (which may be actual AI-2), and second to engineer E. coli to flocculate in response to the presence of C. vulgaris. The first task will be accomplished by transforming a LuxS-mutant strain of E. coli (cannot produce its own AI-2) with an AI-2 reporter biobrick. We will then harvest supernatant from C. vulgaris, which should contain AI-2 or its mimic, and apply it to the reporter strain to test its response. The second task will be accomplished by ligating a gene that causes over-expression of pili (characterized by the pili team) to the Lsr promoter, which is derepressed in response to AI-2. By transforming this part into LuxS-mutant E. coli, we hope to create strain that will stick to algae and will flocculate only in the presence of C. vulgaris.
Virus Surface Display
Oil Sands
References
1. Vetsch, M., Puorger, C., Pilus chaperones represent a new type of protein-folding catalyst. Nature 431, 329-332 (2004).
 "
"

