Team:Michigan/Modeling
From 2010.igem.org
(→Quorum Sensing) |
(→Quorum Sensing) |
||
| Line 33: | Line 33: | ||
Quorum sensing has been modeled by other iGEM teams for previous competitions, including: [http://parts.mit.edu/igem07/index.php/Bangalore Bangalore] and [https://2008.igem.org/Team:NTU-Singapore Singapore] | Quorum sensing has been modeled by other iGEM teams for previous competitions, including: [http://parts.mit.edu/igem07/index.php/Bangalore Bangalore] and [https://2008.igem.org/Team:NTU-Singapore Singapore] | ||
[[Image:LuxS- Modeling001.jpg|right]] This model is based on the paper WILLFINDEVENTUALLYI'MSORRY and was created with the help of [[User:infekt|Alex]] who is also on the quorum sensing team. More information about quorum sensing can be found here: [[Team:Michigan/Project]] and their notebook can be found here: [[Team:Michigan/Quorum_Sensing]]. | [[Image:LuxS- Modeling001.jpg|right]] This model is based on the paper WILLFINDEVENTUALLYI'MSORRY and was created with the help of [[User:infekt|Alex]] who is also on the quorum sensing team. More information about quorum sensing can be found here: [[Team:Michigan/Project]] and their notebook can be found here: [[Team:Michigan/Quorum_Sensing]]. | ||
| + | <br style="clear: both" /> | ||
==Surface Display== | ==Surface Display== | ||
Revision as of 05:31, 18 August 2010
Mathematical models are very important in the design of an iGEM project. The Michigan modeling team is composed of Josh, Kevin, Candy and Jennifer. We are currently researching different models that have been previously done that can help us in our own modeling endeavors. To construct our models, we have taken advantage of MATLAB's Simbiology toolset.
There are several components to a good mathematical model. These include assumptions, parameters, and equations.
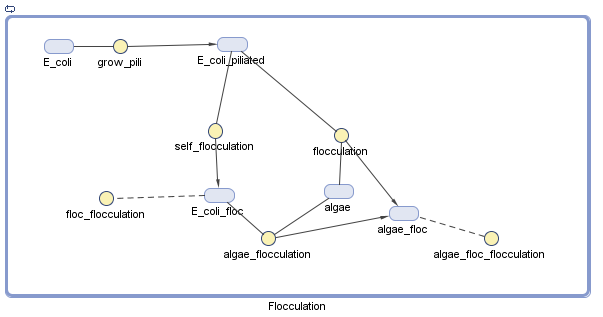
Pili
We have decided to model the growth of a biofilm using pili. By modeling the flocculation of the algae, we hope to be able to predict how our reactions will proceed and determine quantities such as the optimal initial concentration of E. coli.
Assumptions
There is no growth or degradation of cells and algae.
- I made this assumption to simplify the model.
Floc formation only involves two particles. Collision efficiency is perfect.
- This assumption greatly simplifies the model.
Parameters
All rate constants set to 0.1.
- This is obviously unrealistic, but with experimental data, we can refine the numbers.
Initial quantities of E. coli and algae are 1 mol.
- We can perform assays to determine the ideal starting conditions.
The pili neural network has been characterized in several papers [1][2]. Essentially, the two recombinases FimB and FimE control an invertible DNA element that acts as a switch, known as FimS. When FimS is in the "on" position, the cell becomes fimbriated. It has been previously determined that the level of piliation depends on the ratio [FimE]/[FimB]. The goal of the modeling team is to determine the optimal ratio to promote flocculation.
Quorum Sensing
Quorum sensing has been modeled by other iGEM teams for previous competitions, including: Bangalore and Singapore
This model is based on the paper WILLFINDEVENTUALLYI'MSORRY and was created with the help of Alex who is also on the quorum sensing team. More information about quorum sensing can be found here: Team:Michigan/Project and their notebook can be found here: Team:Michigan/Quorum_Sensing.
Surface Display
Oil Sands
We are developing a mathematical model which describes the degradation of naphthenic acids by Pseudomonas putida and Pseudomonas fluorescens in a biofilm.
References
1. Kuwahara, H., Myers, C., Samoilov, M., Abstracted Stochastic Analysis of Type 1 Pili Expression in E. Coli.
2. Wolf, D., and Arkin, A., Fifteen Minutes of fim: Control of Type 1 Pili Expression in E. Coli. OMICS 6 2002
 "
"