Team:Kyoto/Project/Goal A
From 2010.igem.org
(→Introduction) |
(→Method) |
||
| Line 10: | Line 10: | ||
===Method=== | ===Method=== | ||
In order to measure RPU of R0011, we use three types of E.coli, E.coli KRX transformed with <partinfo>BBa_K358000</partinfo>, KRX transformed with <partinfo>BBa_K358001</partinfo>, KRX transformed with <partinfo>pSB4K5</partinfo> without insert. | In order to measure RPU of R0011, we use three types of E.coli, E.coli KRX transformed with <partinfo>BBa_K358000</partinfo>, KRX transformed with <partinfo>BBa_K358001</partinfo>, KRX transformed with <partinfo>pSB4K5</partinfo> without insert. | ||
| + | |||
| + | The reason we used KRX is that lacI repressor is over expressed in KRX and lacI can repress lactose promoter fully at low IPTG concentration <sup>[[#RefA003|[3]]]</sup>. | ||
[[Image:KyotoFigA001.png|300px|left]] | [[Image:KyotoFigA001.png|300px|left]] | ||
| Line 16: | Line 18: | ||
[[Image:KyotoFigA004.png|300px|right]] | [[Image:KyotoFigA004.png|300px|right]] | ||
| - | + | In order to measure RPU, we must use two types of E.coli transformed with the test promoter circuit and the standard promoter circuit respectively to compare absolute activity of test promoter with that of the standard. BBa_K358000 is the lactose promoter, R0011 with GFP and BBa_K358001 is the standard promoter with GFP. Both parts are combined with pSB4K5, a low copy plasmid, because lacI cannot repress lactose promoter enough in low IPTG concentration if there is too much lactose promoter DNA. The E.coli transformed with pSB4K5 without the insert is also used to correct the autofluorescence. | |
| - | RPU is | + | We pick up three colonies from each plate, and cultivate them in the supplemented M9 medium for a night, about 16 hours. The overnight cultures were diluted to 1:100 in pre-warmed fresh the supplemented M9 medium. 3h later and 3.5h later, we measure OD600 of the culture and GFP fluorescence in the culture. We firstly measure RPU after cells are lyzed. The measurement of RPU with some IPTG is done without cell lysis. |
| - | + | To know more detail procedures, go [[Team:Kyoto/Protocols|Protocols]] and [[Team:Kyoto/Notebook2|Notebook]]. | |
===Result=== | ===Result=== | ||
Revision as of 00:43, 27 October 2010
Contents |
Goal A: Characterization of R0011, a strong lactose promoter
Introduction
We used <partinfo>R0011</partinfo> [learn more], a lactose promoter, in the experiment of lysis box.
Because part parameters of R0011 are partially uncertain [1], we characterized R0011 in order to characterize lysis cassette.
We characterize R0011 by Relative Promoter Unit (RPU) [learn more]. When absolute promoter activity is measured at various test condition and measurement instruments, there is variation in promoter activity; RPU can reduce this Coefficient of variation (CV) from 39.1% to17.5% [2]. Therefore RPU can make it easier for us to share the data of promoter activity and use biobricks.
Method
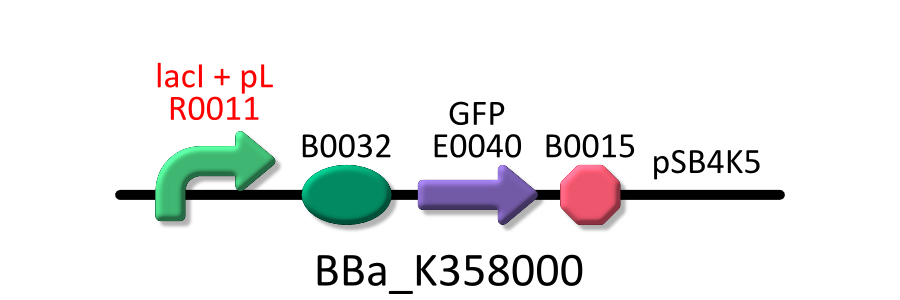
In order to measure RPU of R0011, we use three types of E.coli, E.coli KRX transformed with <partinfo>BBa_K358000</partinfo>, KRX transformed with <partinfo>BBa_K358001</partinfo>, KRX transformed with <partinfo>pSB4K5</partinfo> without insert.
The reason we used KRX is that lacI repressor is over expressed in KRX and lacI can repress lactose promoter fully at low IPTG concentration [3].
In order to measure RPU, we must use two types of E.coli transformed with the test promoter circuit and the standard promoter circuit respectively to compare absolute activity of test promoter with that of the standard. BBa_K358000 is the lactose promoter, R0011 with GFP and BBa_K358001 is the standard promoter with GFP. Both parts are combined with pSB4K5, a low copy plasmid, because lacI cannot repress lactose promoter enough in low IPTG concentration if there is too much lactose promoter DNA. The E.coli transformed with pSB4K5 without the insert is also used to correct the autofluorescence.
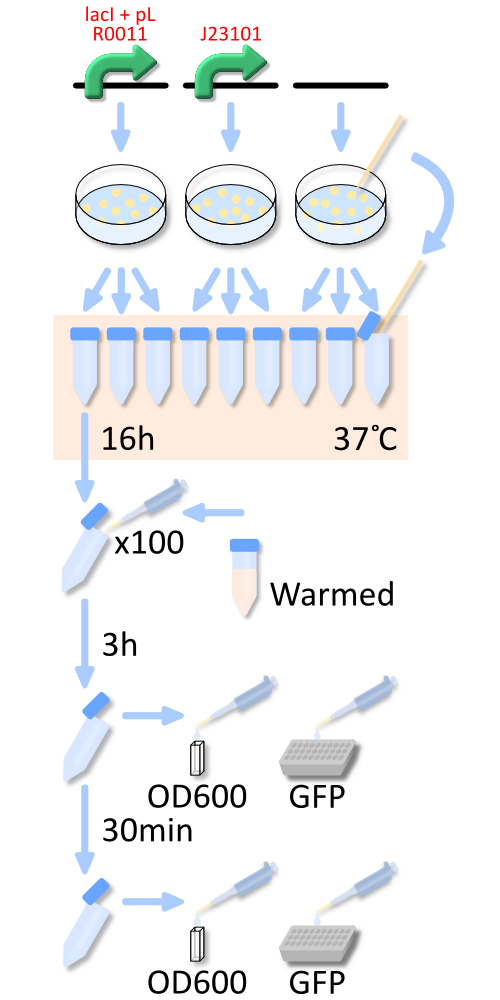
We pick up three colonies from each plate, and cultivate them in the supplemented M9 medium for a night, about 16 hours. The overnight cultures were diluted to 1:100 in pre-warmed fresh the supplemented M9 medium. 3h later and 3.5h later, we measure OD600 of the culture and GFP fluorescence in the culture. We firstly measure RPU after cells are lyzed. The measurement of RPU with some IPTG is done without cell lysis.
To know more detail procedures, go Protocols and Notebook.
Result
The result of characterization is shown in table and figure below.
| IPTG (mM) | RPU | |||||
|---|---|---|---|---|---|---|
| A | B | C | Average | Stdev | CV | |
| 0.01 | 0.0191 | 0.00963 | 0.00244 | 0.0104 | 0.0084 | 0.806 |
| 0.03 | 0.177 | 0.155 | 0.198 | 0.174 | 0.0173 | 0.0997 |
| 0.1 | 0.205 | 0.191 | 0.269 | 0.221 | 0.0416 | 0.188 |
| 0.3 | 0.938 | 0.955 | 0.955 | 0.949 | 0.00981 | 0.0103 |
| 0.5 | 0.263 | 0.354 | 0.408 | 0.342 | 0.0733 | 0.215 |
| 0.8 | 1.44 | * | 1.21 | 1.37 | 0.161 | 0.122 |
| 1.0 | 1.60 | 1.58 | 1.65 | 1.65 | 0.106 | 0.0640 |
| 2.0 | 1.49 | 1.83 | 1.39 | 1.57 | 0.233 | 0.148 |
- *One of RPU in IPTG 0.8mM couldn’t measured.
Modeling
We make the model about the activity of R0011 and decide parameters from the result. The result is also drawn in figure above. To know more about this model, see Modeling.
Discussion
The result indicates that the maximum activity of R0011 is about 1.6 RPU and the minimum is less than 0.01 RPU and R0011 has wide range of activity (maximum activity of R0011 is about 160 fold greater than the minimum activity at least). The wide range of activity of RPU can change the effect of lysis cassette largely, and it is useful to characterize lysis cassette. CV in 0.01mM IPTG is 0.806, so large, we cannot trust RPU in 0.01mM ITPG so much.
 "
"