Team:Kyoto/Project
From 2010.igem.org
Contents |
Project
Introduction
Background
The cell-death device is required in many fields[1,2]. In bioremediation, for example, this device can prevent introduced bacteria from disrupting the ecosystem[3,2HP]. Also, It can be applied to other ideas such as E.coli capsules which scatter drug or aroma by cell-lysis responding to a certain signal. Therefore, the cell death devices based on diverse approaches, including previous iGEM projects have been developed[1,3,4,5HP]. However, present cell-death devices are still not easy to use, because of the poor characterization or strict restriction on promoters' activity and so on[6,7HP]. Therefore, we attempt to register on BioBrick better characterized and more useful cell-death device to contribute to every future team.
Problems of Present Cell-Death Devices
In order to design a universal and applicable cell-death device, we need to reduce the restriction on promoter's activity and to characterize the strength of the cell-death function.
1. Need for Characterization
In fact, there is no experimental data about the present devices in the iGEM's cell-death category. Quantitative analysis of Lysis cassette is necessary. To contribute to iGEM in this category with well characterized device, we did experimentation and some modeling.
2. Solution to the Restriction on Promoter's Activity
In constructing a simple circuit consisting of an inducible promoter and a killer-gene, the combinations are limited by the promoter's leak, strength of killer-function and many other factors. We looked for the solution to this problem and came up with an idea of using an anti-killer gene as well with two promoters (a constitutive one and an inducible one). With this circuit, we can apply more promoters to this device by using an appropriate constitutive promoter.
We considered that Lysis cassette of λ phage and its antagonist, SΔTMD1 are the most appropriate to Killer-gene and Anti-killer gene[4]. This was because some iGEM teams in the past already dealt with lysis cassette and also because we found some articles that shows the effect of SΔTMD1 in the yeast[10].
Learn more about Lysis Cassette.
Overview
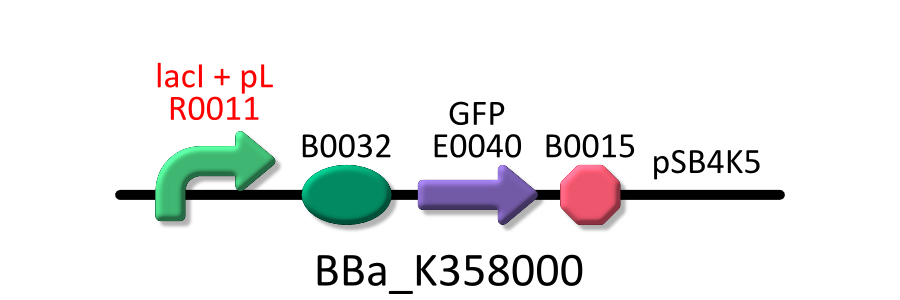
Goal A: Characterization of R0011, a strong lactose promoter
We used R0011, which is one of the most commonly used parts in iGEM, to characterize our genes because the strength of promoter activity is very high. However, in previous iGEM, no teams characterized R0011 with RPU. We tried to determine what is the ratio of activity of R0011[9].
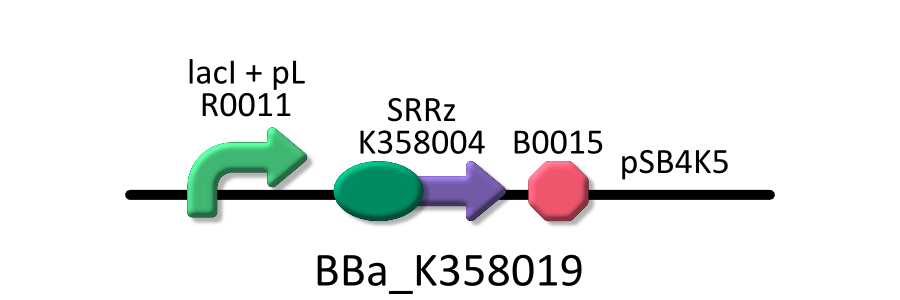
Goal B: Characterization of λ Lysis cassette
To characterize λ Lysis cassette, we cultivated E.coli transformed with the device in various concentration of IPTG. Then, we measured A550 of the cultures, and determined the lytic activity of λ Lysis cassette depending on the activity of its promoter.
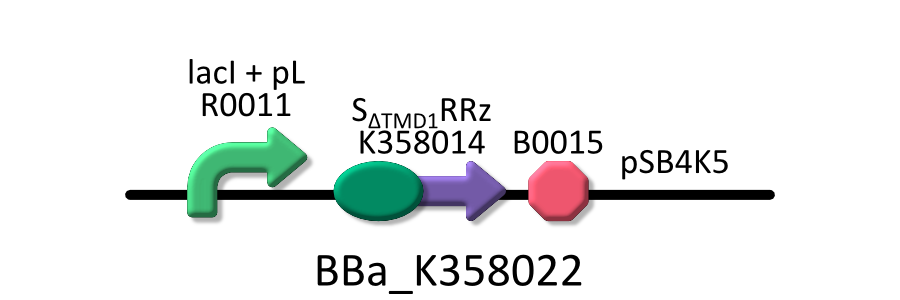
Goal C: Characterization of the anti-killer gene, SΔTMD1
We made upper construct and measured A550 of E.coli transformed with the device in medium with IPTG and without IPTG[5,6,7,8].
Conclusion
1 We measured RPU of R0011 in eight kinds of concentration of IPTG, and made c 2 We made a quantitative measurement standard of lytic activity of λ Lysis cassette. 3 We characterized λ Lysis cassette quantitatively, and found out a new fact that cell lysis become an equilibrium state depending on the strength of induction(RPU). 4 We checked the activity of SΔTMD1、which can't form pore of holin and cells don't die although endolyshin expresses correctry.
Future work
Methods
See pages below for details.
- Protocols: Protocols of each experiment such as Polymerase Chain Reaction (PCR), Restriction Digestion, Ligation, Transformation.
- Materials: Strains, DNA, and Primers.
- Parts: Construction of each part and BioBrick Parts used in our project.
- Notebook: Laboratory notebook.
- Team:Kyoto/LearnMore: <一時的においています。>
 "
"