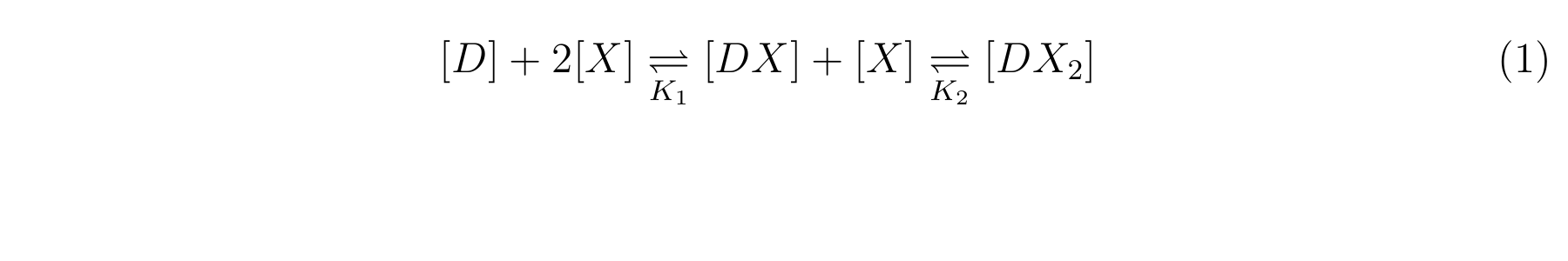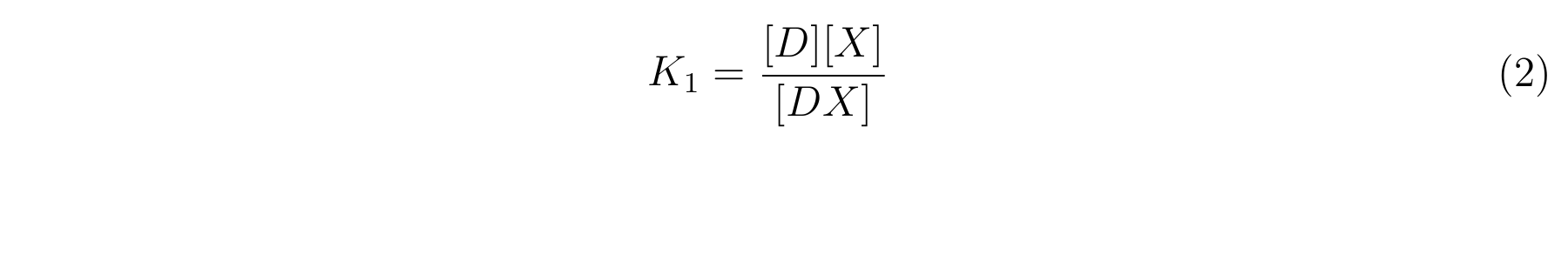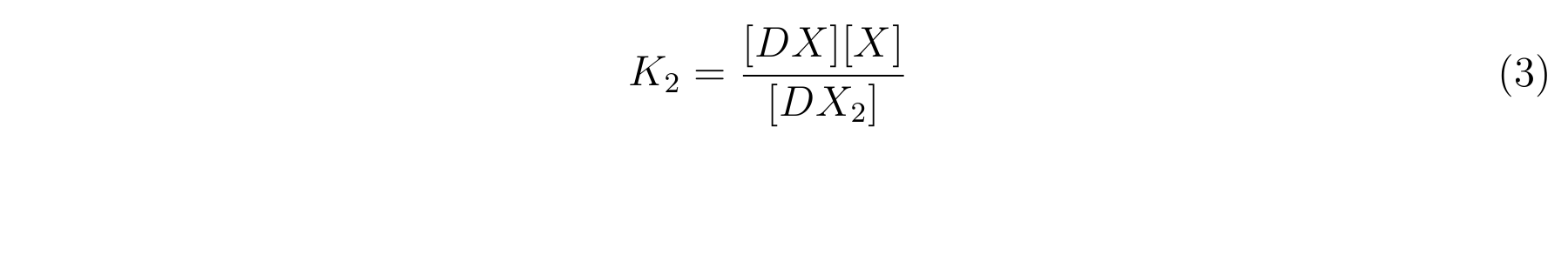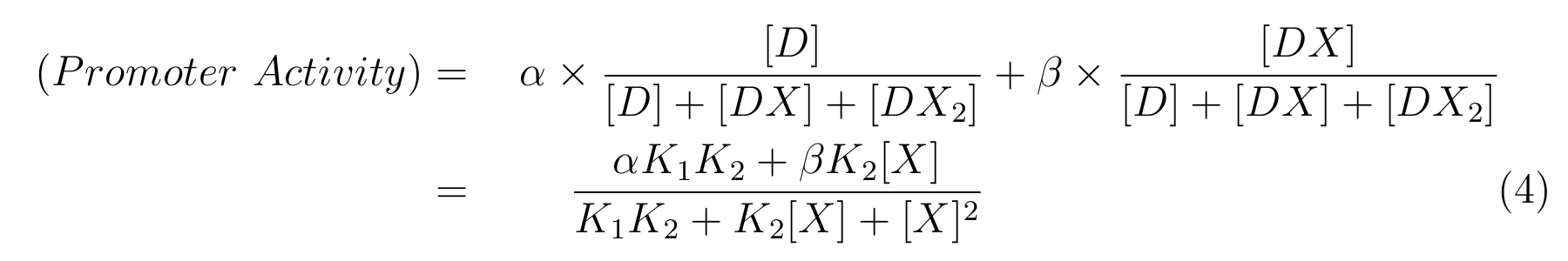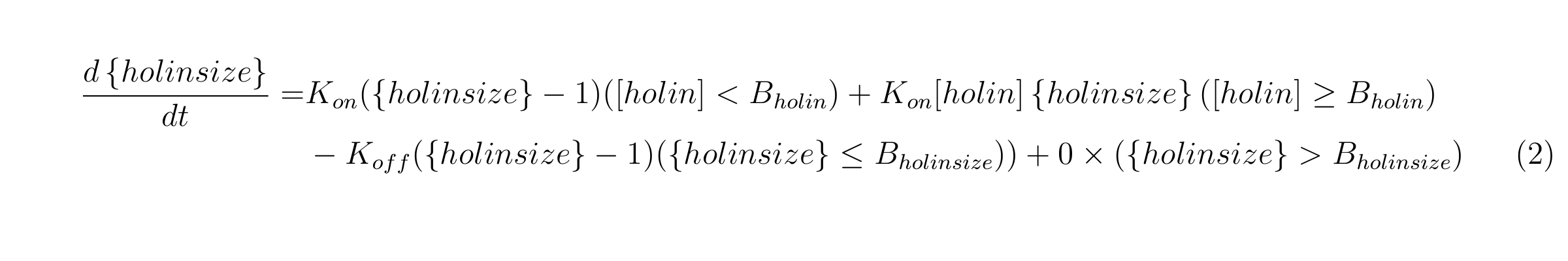Team:Kyoto/Modeling
From 2010.igem.org
Contents |
Modeling
Models
Model1: R0011, a lactose promoter
Before making the model for lysis cassette, we must characterize R0011, a lactose promtoer, because we use it to change the expression level of lysis cassette. We made some mathematical model for the lcatose promoter activity.
Model2: Cell Lysis
Model1: R0011, a lactose promoter
Indroduction
It is too laborious to measure the activity of R0011 with all IPTG concentrations, such as 0.01mM, 0.02mM, , , , , 10mM. Measurement with some appropriate IPTG concentrations and making a mathematical model from the experimental data is beneficial to estimate the promoter activity with the other IPTG concentration and save the trouble.
LacI binds to lactose promoter and represses it
figure
LacI, the repressor of lactose promoter, binds to DNA sequence of lactose promoter and represses it. R0011, a lactose repressor, has two operator regions where LacI binds [1]. The equilibrium reaction of binding and dissociation of LacI and the lactose promoter can be described as equation1.
Here,
- [X] means the concentration of LacI,
- [D] means the concentration of the lactose promoter not binding LacI,
- [DX] means the concentration of the lactose promoter binding a lacI
- [DX2] means the concentration of the lactose promoter binding two LacI.
- K1 means the equilibrium constant for the reaction between [D] and [X],
- K2 means the equilibrium constant for the reaction between [DX] and [X].
- The equilibrium constants K1 and K2 are described as equation2 and equation3 respectively.
From equation2 and equation3, equation4 is established.
Here,
- α is the promoter activity when all the lactose promoter bind two LacI respectively,
- β is the promoter activity when all the lactose promoter bind a LacI respectively,
-
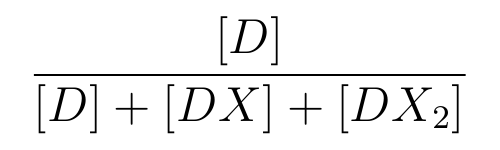 is the ratio of [D] to the total concentration of lactose promoter,
is the ratio of [D] to the total concentration of lactose promoter,
- 100px is the ratio of [DX] to the total concentration of lactose promoter.
In this model, we hypothesized that the promoter activity is 0 when all the lactose promoter bind two LacI respectively.
LacI binds to lactose promoter and represses it
figure
The inducer of lactose promoter, Lactose and IPTG, binds to LacI, and changes LacI conformation so that LacI dissociates from lactose promoter. The equilibrium reaction of the inducer and LacI is described as follows.
equation5
Here, [SX] is the inducer concentration, [X] is the concentration of LacI not binding with the inducer, n is the number of the inducer molecule binds to one LacI molecule. The equbilium constant, KXn, is described as equation6.
equation6
We assume that the total concentration of LacI in a cell does not change in the log phase growth in which the cell growth does not change because the dilution effect for LacI due to the cell growth does not change and the expression and the degradation of LacI is constitutive. Therefore, let XT as the concentration of total LacI, and equation7 is applied.
equation7
From equation6 and equation7, equation8 is established [2].
equation8
Lactose and IPTG are inducer of lactose promoter
figure
From equation4 and equation8, the relationship between IPTG and lactose promoter activity is described as follows.
equation9
Simulation
We apply equation9 to the result of characterization of R0011 and parameters are decided by using MATLAB 7.10.0 (MathWorks).
graph
The orange markers and error bars are experimental data and blue line is the expectation from this model. The data of 0.5mM IPTG does not be used in fitting the model.
Parameters are shown in table below.
table
Reference
[1] http://partsregistry.org/Part:BBa_R0011
[2] Uri Alon (2006) “An Introduction toSystems Biology” Chapman & Hall
Model2: Cell Lysis
Assumptions
It is known that when lambda phage holin is expressed, they localized to plasma membrane.
Tough, holin in the plasma membrane is monomer because there is a few holins there firstly, holin becomes oligomer when enough holins exist in the plasma membrane. It is suggested that if the number of holin in the oligomer reaches about 62, holin oligomer makes a hole [1]. Once the hole is made, endolysin can go to the periplasm, and degrades peptideglycan and make cell lysis.
In order to make a model that can explain our experimental data for cell lysis, we made some assumptions.
The first is that there is limit of the number of holin in the plasma membrane. We think this is because the surface area of plasma membranes is limited. It is estimated that approximately 1000-3000 molecules of holin accumulates in the plasma membrane finally [2].
Next one is that when only a few holin exist in the plasma membrane, the speed of binding between holin molecules is so low, but the speed is increased when the number of holin molecule becomes larger.
The next is that the size of holin oligomer is limited and that there is 62 holin molecules in the holin oligomer at most.
We also hypothesized that the effect of endolysin is neglected while the number of holin molecules in the ologomer is under 62, but endolysin works when it reaches 62 and that the effect of endolysin becomes larager as the number of endolysin molecules increase, but there is limit for the effect of endolysin.
Equations
We made equations below.
equation3 equation4
Equation1
equation1
In this equation, [holin] means the concentration of holin in the plasma membrane, Aholin is the constant including the transcriptionarate ,the translational rate and the rate of transport to the plasma membrane, γholin means the dilution and degradation rate of holin.
is the promoter activity of lactose promoter. [IPTG] is [Sx] in the eqaution9 of “Model for the activity of lactose promoter, R0011”. To know more, go Model for the activity of lactose promoter, R0011
We also assume that although while [holin]<=3000, [holin] is increasing, [holin] is in steady state when [holin]>3000.
Equation2
equation2
{holin size} means the number of holin molecules in holin oligomer, Kholin_on is the binding speed of holin monomers or oligomers while [holin]<Bholin, Kholin_size’ is the binding speed of holin molecules when [holin] >= Bholin. Bholin is the thresh hold for the binding speed of holin molecules begin to increase. Kholin_off is the dissociation speed of holin oligomers. We also assume that once {holin size} reaches Bholin, holin oligomer make a hole and {holin size} does not change.
Equation3
equation3
In this equation, [endolysin] is the concentration of endolysin, Aednolysin is the constant including the transcriptional rate and the translational rate of endolysin, γendolysin is the dilution and degradation rate of endolysin.
is the lactose promoter activity.
Equation4
equation4
In this equation [cell] is the number of cells in the medium, r is the cell growth rate, Kcell is the carring capacity, D is the constant for cell lysis Kendolyisn is the threshold of the effect of endolysin for cell lysis.
We assume that while {holin size}<62, holin does not make a hole and endolysin does not degrade peptideglycan and the effect of endolysin is 0. We also assume that after holin molecules make holes, though, the effect of endolysin increases as the concentration of [endolysin] increases, there is limit for the effect of endolysin.
Simulation
Reference
- Ry Young et al. “The holin of bacteriophage lambda forms rings with large diameter” Molecular Microbiology (2008) 69(4), 784–793
- Chang, C.Y., Nam, K., and Young, R. (1995) “S gene expression and the timing of lysis by bacteriophage l.” J Bacteriol 177: 3283–3294.
 "
"