Team:Imperial College London/Results
From 2010.igem.org
| Line 178: | Line 178: | ||
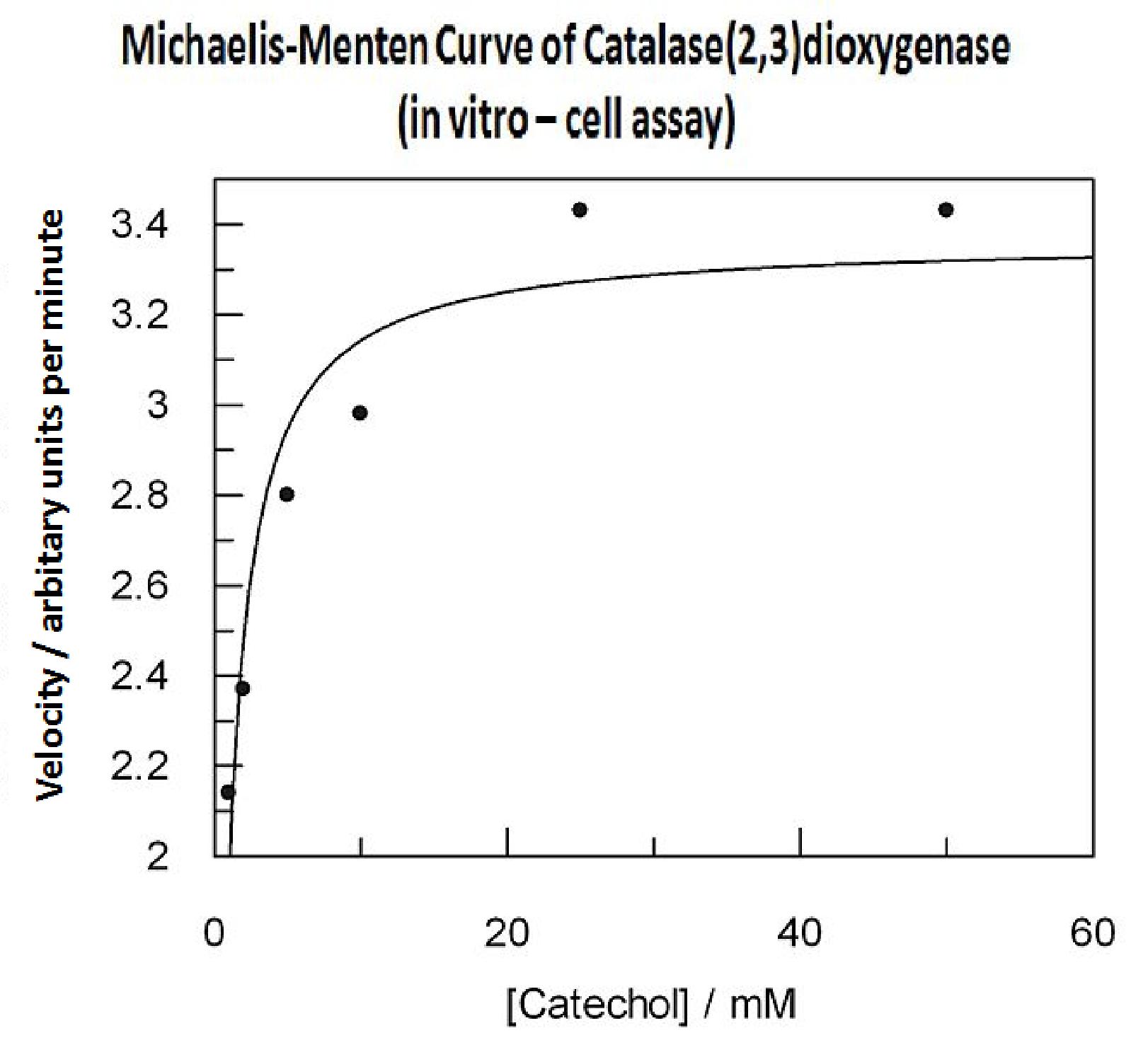
Data collected from the assay allowed us to construct a Michaelis-Menten curve for the in vitro kinetics of Catechol(2,3)dioxygenase in cell lysate. The cell lysate was obtained from a 100ml overnight culture and the lysate was diluted by a factor of 20-fold in order to obtain a suitable concentration of total enzyme for the plate reader assay. The concentrations of catechol used to derive Michaelis-Menten curve were 1, 2, 5, 10, 25, 50 mM, prepared from 1M stock catechol. | Data collected from the assay allowed us to construct a Michaelis-Menten curve for the in vitro kinetics of Catechol(2,3)dioxygenase in cell lysate. The cell lysate was obtained from a 100ml overnight culture and the lysate was diluted by a factor of 20-fold in order to obtain a suitable concentration of total enzyme for the plate reader assay. The concentrations of catechol used to derive Michaelis-Menten curve were 1, 2, 5, 10, 25, 50 mM, prepared from 1M stock catechol. | ||
| - | + | [[Image:Grapfit curve 1.jpg]] | |
|} | |} | ||
Revision as of 21:56, 26 October 2010
| Results |
| C230 |
|
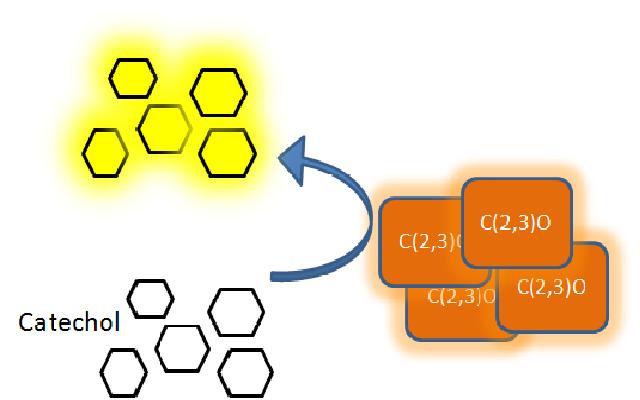
The XylE gene, originating from the TOL plasmid of Pseudomonas putida, was the choice for the output signal mediator of our system. The system is based on catechol 2,3-dioxygenase enzyme. Colonies of cells that express xylE gene product become yellow within seconds after selection plates are sprayed with catechol, a colorless substrate, that is converted by CatO2ase to the yellow product, 2-hydroxymuconic semialdehyde. This reporter system is ideal for our project as it has very fast kinetics, gives a visual output by naked eye and the substrate used is very cheap. |
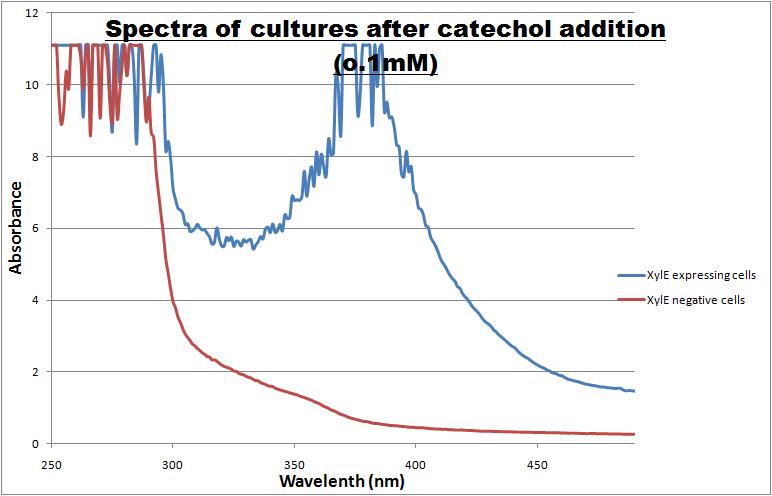
| Experiment 1 | Optimum absorption wavelength for catechol assays |
|
The CatO2ase protein, except as an ideal output signal for our engineered bacterial detector it can also serve as a great reporter gene. Its characteristics can be exploited in a wide range of fields across biological sciences field, so it was one of the team's first candidates for further characterization. Qualitative and quantitative data were gathered and presented. All further tests involving XylE transformed cells were quantified by measuring absorbonce at 380nm wavelenth. Catechol ,the initial substrate of CatO2las enzyme, is colourless. However within seconds of its addition the colonies/liquid cultures of XylE expressing cells become yellow, indicating production of product which absorbs light in the region visible spectrum. An spectrophotometric assay was prepared, where the spectra of two cultures of E.coli (one XylE gene transformed and the other not)were compared on addition of 0.1mM Catechol substrate. The spectra (figure 1) showed that in XylE transformed cells, a broad peak appears at about 380nm. The absorbance of the particular wavelengths of light by the product of the enzymatic reaction 2-hydroxymuconic semialdehyde, is what causes the yellow output.
References
|
| Experiment 2 | The threshold value of catechol assay |
|
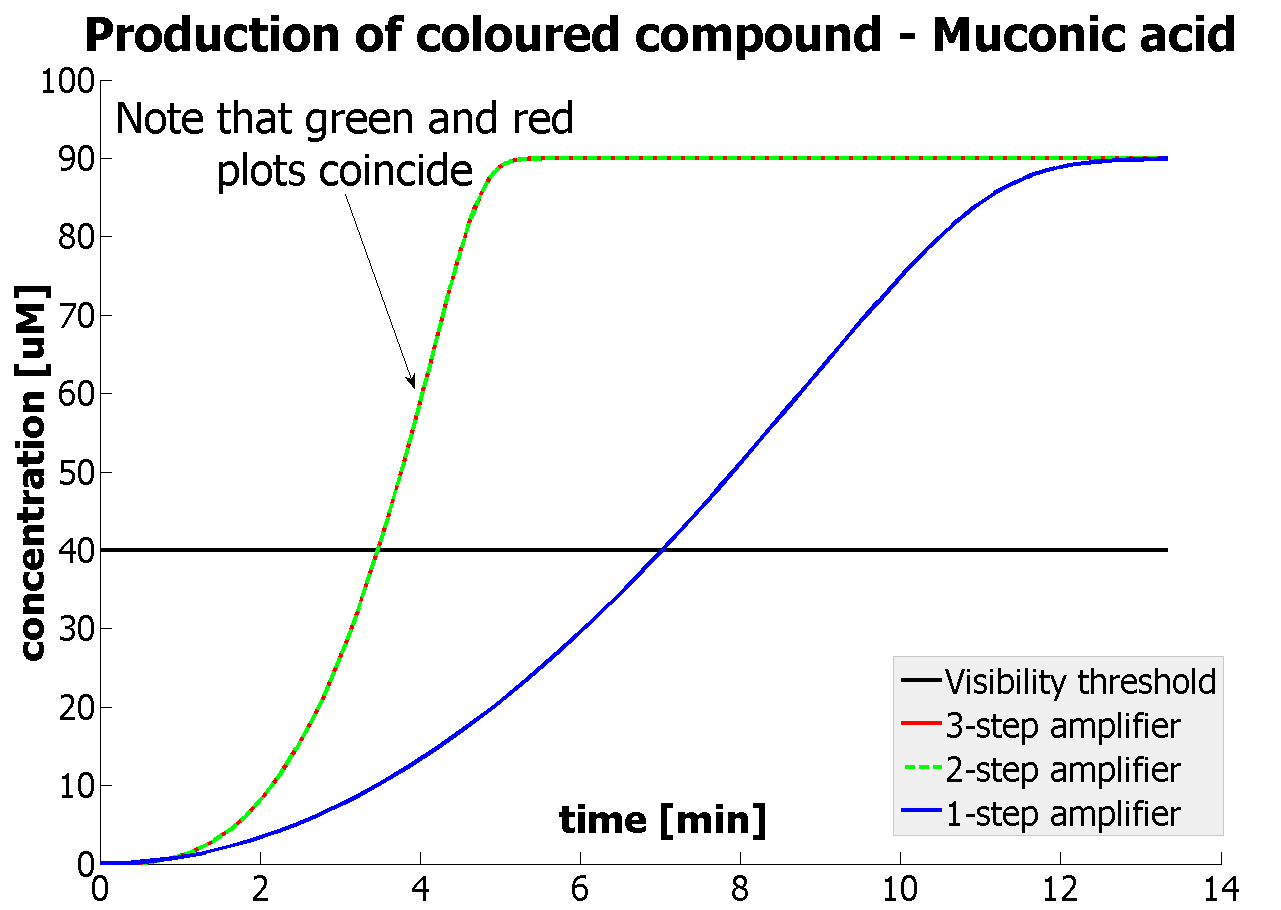
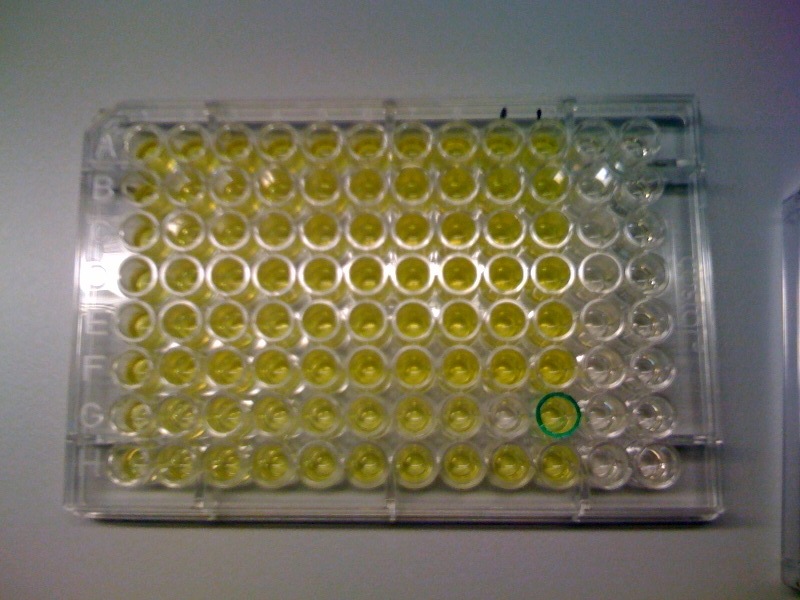
Addition of catechol to a a culture of cells constitutively expressing C(2,3)0 enzyme results into production of yellow colonies on plate or yellow liquid cultures if E.coli is grown in liquid medium. In a lab where a spectrophotometer is readily available anyone can accurately quantify production of even the smallest quantity of the yellow 2-hydroxymuconic semialdehyde product. However, as our biological detector is intended for field use where sophisticated spectroscopic equipment is not available, it was obvious that we needed to establish the threshold value at which a positive result (yellow color) would be detectable by the naked eye. The set up of this experiment involved preparation of XylE expressing E.coli cultures at an O.D. of 0.5 in LB and M9 medium. Then a gradient of catechol concentrations was prepared and added to the cell cultures. A group of 5 individuals, but not the lab member preparing the test samples, were given the test cultures and asked where on the gradient they were not able anymore to see the yellow color. The 2 concentrations where the transition from yellow to colorless occurred were identified and a second gradient of catechol concentrations was prepared. In this second gradient the gap between the 2 concentration was analysed. The highest catechol concentration was the least yellow from the first part of the experiment, while the lowest catechol concentration was the first "colourless" concentration from the first part of the experiment. Again the group of 5 testers identified the transition concentrations from yellow to colorless. The threshold concentration of catechol needed for a person to identify a yellow positive test was found to be 30-40μM. This result was fed back to the modelling team and allowed them to constrain the concentration values of catechol to be used. Furthermore, it facilitated the determination of conditions for 2-step amplification performing better than 1-step amplifier. If you want to know more on output amplification modelling, please follow the link. |
| Experiment 3 | Characterizing kinetic parameters of C23O in whole cells |
|
In our Parasight project the XylE gene product, C(2,3)lase enzyme, is the means by which an output signal is generated. If the parasite is present is the water supply, the bacterial system is activated by an input signal which produces the yellow color, the positive output signal. Test assays in the lab on colonies of cells expressing XylE gene, become yellow only seconds after addition of catechol (100mM). From this it was realized our group that the kinetics of the C(2,3)lase enzyme are such that make it an attractive candidate as a reporter enzyme. So, we decided that further characterization of the kinetics of the C(2,3)lase might help identify a really efficient reporter for research centers.
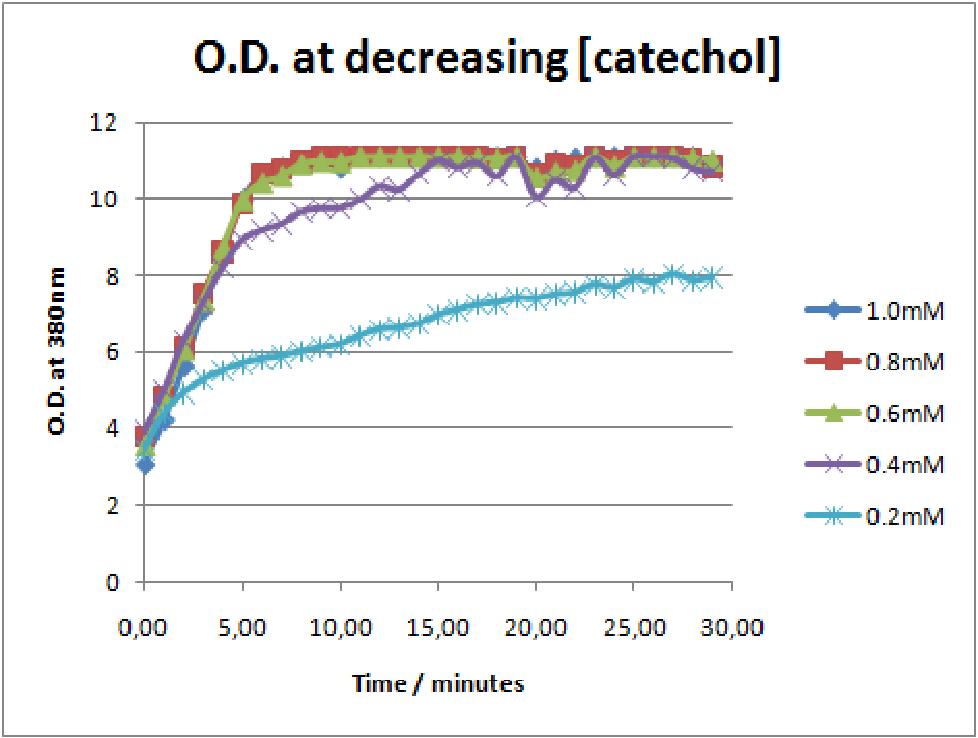
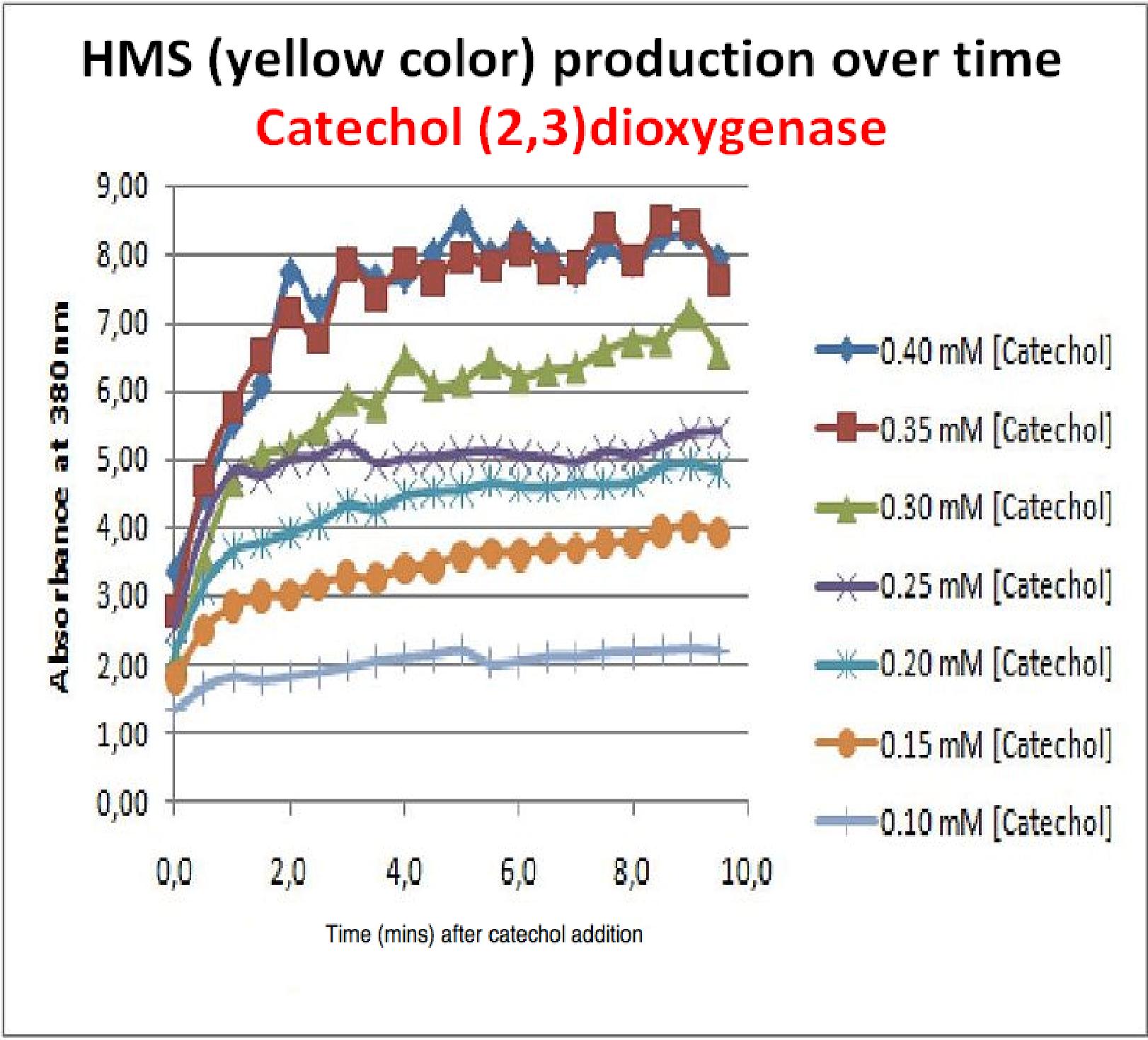
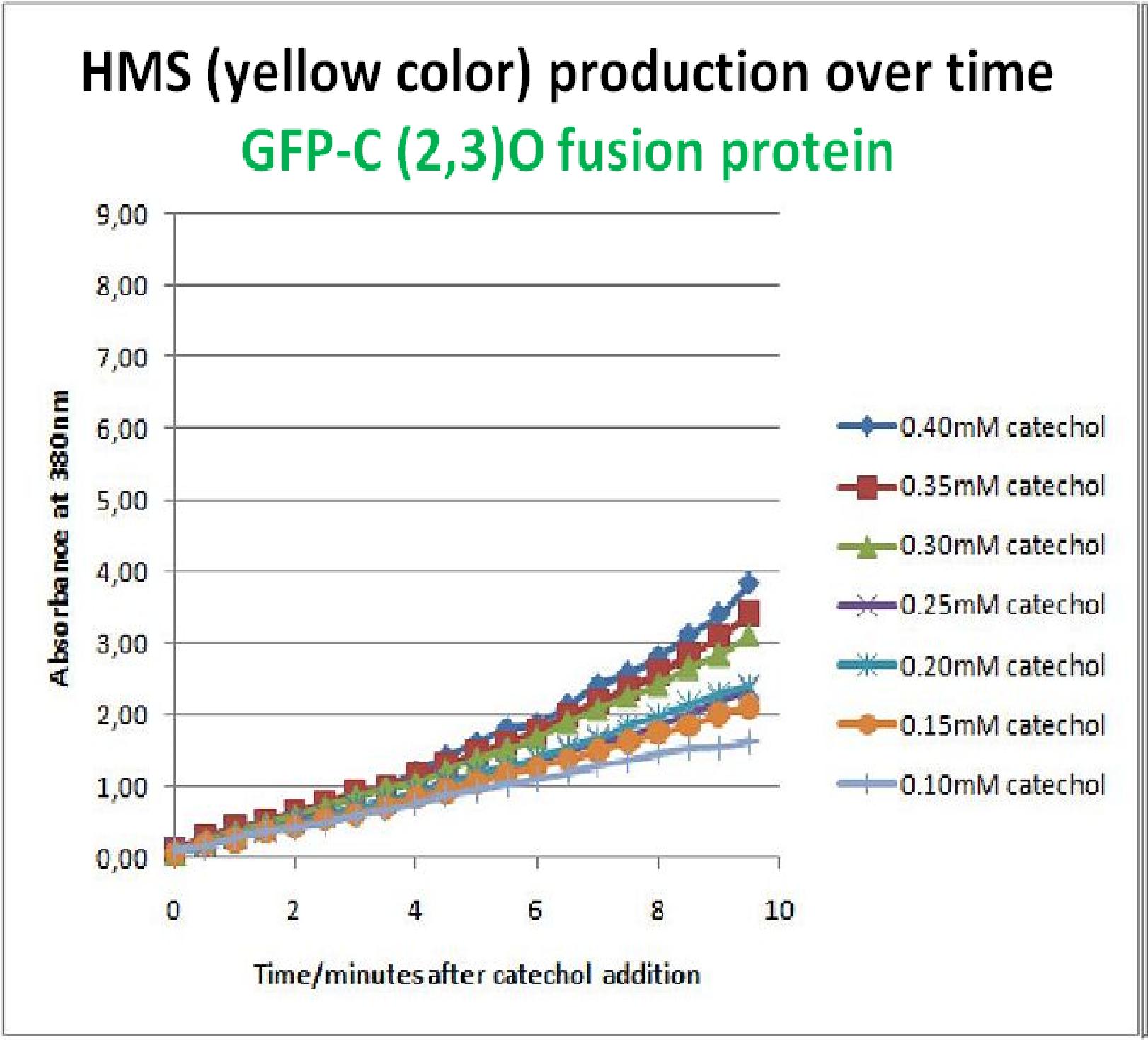
The results of the experiment confirmed previous hints of the huge catalytic turnover rate of the C(2,3)0 enzyme. The graph on the left illustrates production of yellow product per min. The slope at the initial stages of the reaction (first minutes) would represent the rate of the reaction, that is the initial Velocity. By calculating initial velocities of the reaction at different substrate concentrations, one can plot a Michaelis-Menten curve to extract kinetic parameters such as Vmax, Km and catalytic turnover number.
Several attempts were made to determine these parameters on whole cells. Unfortunately due to the actual nature of the enzyme, velocity of reaction and technical limitations of lab equipement this has been proven impossible. To start with one of the limitation was that the color output and the extinction coefficient of the yellow product is so large, that the system saturates within the first minutes after catechol addition to cell cultures. The spectrophotometer and plate reader reaches its detection maximum reading value from the start of the reaction. Also due to the high velocity of the reaction, in the time frame of about 10 seconds that are needed to load the plate into the plate reader after catechol addition, the reaction has already proceeded too far. So initial velocity measurements of the reactions needed for construction of Michaelis-Menten curve are inaccurate. These problem could be overcome by decreasing total enzyme concentration. In whole cell cultures this would be possible by decreasing cell density in each well as each cell contain an average C(2,3)O enzyme concentration. However, previous experiments showed that cell cultures with optical densities lower than 0.5 O.D. introduce serious perturbations in the readings of the spectrophotometer and plate reader detectors. Despite these limitations some useful data were extracted out of these assays. More specifically we acquired data that delineate the course of the reaction in terms of yellow product production over time at various catechol concentrations. The results of one of these assays is presented in the figure below. In order to extract data that will allow characterization of the kinetic parameters of catechol dioxygenase enzyme our lab team proposes purification of the protein from cell lysate, several fold dilution, and in vitro characterization. |
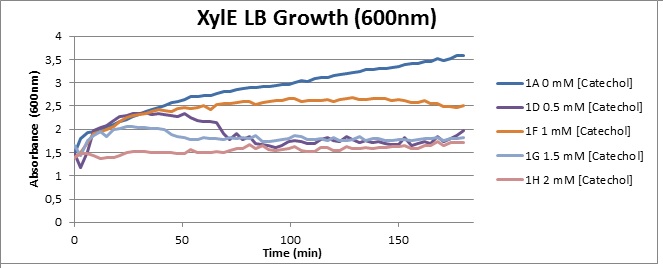
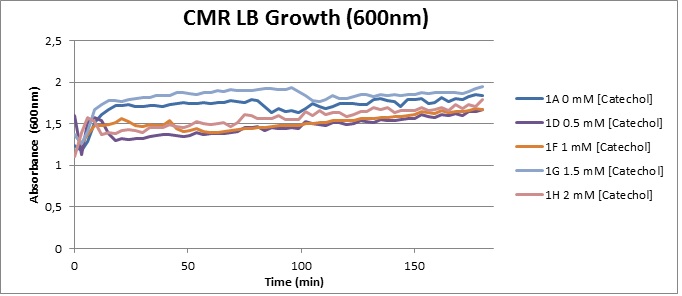
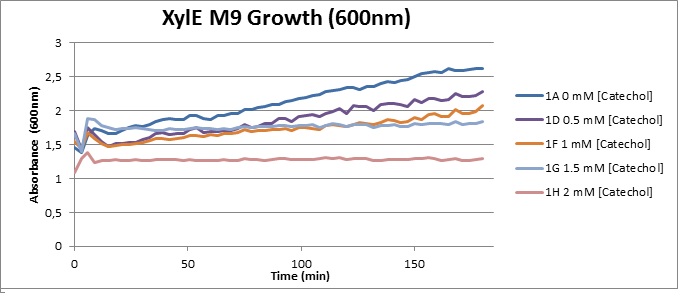
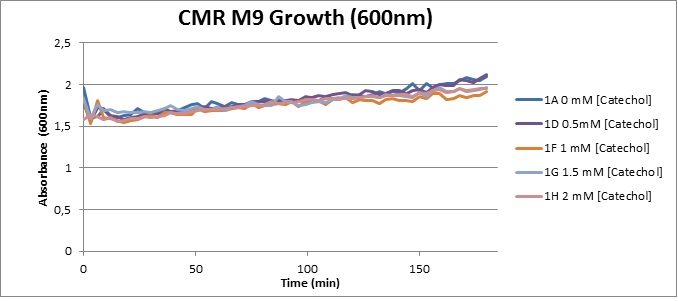
| Experiment 4 | Assaying cell-growth in presence of Catechol | |
|
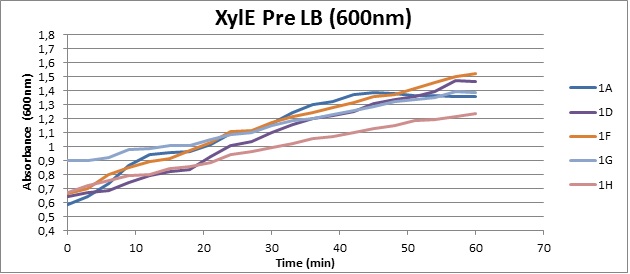
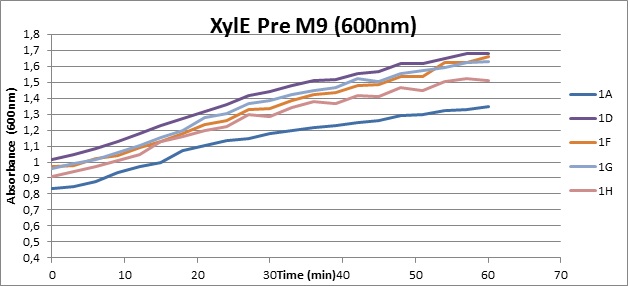
Assaying the growth behaviour of E.coli Top10 cells transformed with either the XylE gene promoted by Pveg or alternatively as control, transformed with a CMR vector of identical size, we were able to determine if and what effect either Catechol itself, or the breakdown product 2-hydroxymuconic semialdehyde had the expected growth-inhibiting effects on the cells. Top10 cells were transformed with XylE and CMR respectively contained on the PSB1C3 vector.Experiments were performed both in LB and M9 medium. The 100ml wells were prepared to display an array of 0 to 2mM [Catechol] exposure. To determine cell-growth behavior we measured absorbance at 600nm using a POLARstar Omega plate-reader set to take measurements every 3 minutes over one hour, without catechol. Catechol was added to all wells after the pre-growth assay was completed and measurements of optical density were taken in 3 minute intervals over 3 hours. Growth without Catechol Pre-growth assays showed growth behavior as expected for cells growing normally. | |
|
Growth with Catechol (LB) The addition of catechol had distinctive effects on the XylE expressing cells growing in LB medium. While at 0% catechol growth-behavior did not show a significant change (dark blue), even the lowest concentration of 0.25% catechol appeared to drastically reduce cell-survival (red). In contrast, CMR-control cells did not change their growing behavior in the presence of catechol. | |
|
From this we conclude that in LB medium, the breakdown product of catechol, 2-hydroxymuconic semialdehyde, has a lethal effect on E. coli. (M9) Cells growing in M9 medium appeared more resistant to the effects of catechol. Even though absorbance at 380 nm increased significantly in well containing XylE expressing cells, indicating strong turnover of Catechol by C23O (4), Catechol did not appear to influence growing behavior in generalizable fashion: while a significant increase in the variance in the sample could be determined. Such was not observed for CMR expressing cells exposed to the same conditions. However, we were not able to establish a clear trend of growing-behavior in the presence of catechol as in case of the LB – XylE samples. | |
|
We conclude that the breakdown product of Catechol has a strong, deleterious effect on XylE expressing cells in an M9 medium, however, cells appear more able to resist the effects of 2-hydroxymuconic semialdehyde in M9 medium. These results support the hypothesis that the breakdown product and not catechol itself has an intense deleterious effect on cell survival. This contributes to the safety of our system, as the output module is inherently coupled to the introduction to a substance lethal to the cells. | |
| Experiment 5 | Characterizing GFP-XylE gene product in whole cell cultures |
|
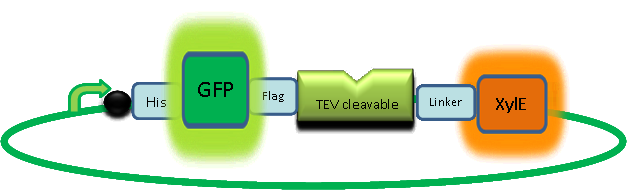
As part of the part of the project, the team needed to identify a reporter gene that would give a visual output. However the means of getting this visual output should be inducible and not constitutively active. Another specification for this reporter was that it was preferred for its activation not to go through the usual route of transcription/translation which is a slow process. We had little success of finding something suitable for our project so we decided to build one. We decided to use the gene product of the XylE gene and by looking at three dimensional diagrams of Catechol(2,3)dioxygenase enzyme and using software tools we proposed an inducible model of this enzyme. In our construct, GFP was added to the N-terminal of C(2,3) monomer. C(2,3) enzyme is a homotetramer were all 4 subunits are required for its activity. We hoped that by adding GFP at the N-terminus of the catechol dioxygenase enzyme, we would block tetramerization of the enzyme, thus render it inactive. This modification is important as it makes our system inducible. The cleavage of the GFP by TEV protease, on arrival of input signal, means that Catechol dioxygenase becomes active only when the parasite is detected. To investigate this, an experiment was designed where catechol was added to GFP-XylE transformed E.coli. The progress of the reaction was monitored by monitoring production of 2-hydroxymuconic semialdehyde (yellow) product using spectrophotometric techniques. This correlates well to the velocity of the reaction, if divided by time, and by using various initial catechol concentrations as a substrate a kinetic profile of the reaction could be constructed. The experimental set up is essentially the same as described above for determining the kinetics of C(2,3)O enzyme. The only difference is that E.coli cells used are the ones transformed with GFP-XylE gene construct instead of XylE gene.
If we take 3 minutes as the reference point on the graphs, as this is where the XylE expressing cells saturate the system with the colored product, we can see that there is about a 10-fold reduction in the rate at which the protein product of GFP-XylE gene construct catalyses the reaction in comparison to the wild type XylE gene. From this we can deduce that the attached GFP protein influences tetramerisation of the Catechol dioxygenase enzyme, so that the equilibrium: 4 monomers (inactive) <-----> tetramer (active) is shifted is shifted towards protein remaining as monomers and thus inactive. The results of this experiment were really encouraging for the team, as it shows that we have managed to inactivate the normally constitutively active reporter protein of our output module. The lab team now proposes an experiment where the inactive fusion protein (GFP-catechol dioxygenase) is exposed to TEV protease. Reversion of the reaction profile back to the kinetics of the wild type Catechol dioxygenase would mean that we have successfully constructed an inducible reporter system. |
 "
"