Team:HokkaidoU Japan/Notebook/September15
From 2010.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
== Result of yesterdays transformation == | == Result of yesterdays transformation == | ||
| - | Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3]. | + | <nowiki> Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3].</nowiki> |
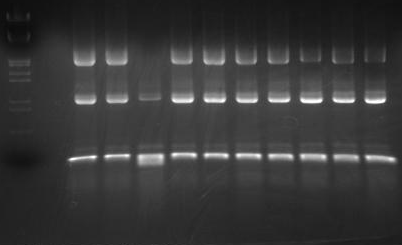
| - | [[Image:HokkaidoU_Pictures_Colony_PCR_of_2010_09_15.png|frame|Colony PCR]] | + | [[Image:HokkaidoU_Pictures_Colony_PCR_of_2010_09_15.png|frame|Colony PCR]]<nowiki> |
| - | Combined length of insert with prefix, suffix and scar is estimated to be 2143bp. From electrophoresis picture under UV light you can see all colonies except colony-3 have bands ant approximate 2000bp position. We used DNA marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png Lamda/(Hindi3, EcoR1)] followings are empty lane and then colonies 1 through 10 accordingly. | + | Combined length of insert with prefix, suffix and scar is estimated to be 2143bp. From electrophoresis picture under UV light you can see all colonies except colony-3 have bands ant approximate 2000bp position. We used DNA marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png Lamda/(Hindi3, EcoR1)] followings are empty lane and then colonies 1 through 10 accordingly. |
We used 1.5% agarose gel for electrophoresis. | We used 1.5% agarose gel for electrophoresis. | ||
| + | </nowiki> | ||
Revision as of 13:09, 15 September 2010
- Observed results of yesterdays transformation
- Transformation using heat shock went well
- Electroporation transformation failed produce colonies
- Did Colony PCR of yesterdays transformed colonies
- Introduced colonies to L(+)Arabinose medium to check if it would show desired function
- Check for results tomorrow
Result of yesterdays transformation
Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3].
Combined length of insert with prefix, suffix and scar is estimated to be 2143bp. From electrophoresis picture under UV light you can see all colonies except colony-3 have bands ant approximate 2000bp position. We used DNA marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png Lamda/(Hindi3, EcoR1)] followings are empty lane and then colonies 1 through 10 accordingly. We used 1.5% agarose gel for electrophoresis. "
"