Team:Groningen/1 June 2010
From 2010.igem.org
| Line 73: | Line 73: | ||
Good bands. The 5’of the primers were designed with BamHI sites for easy circularization of the plasmid. The PCR product was cleaned (High Pure PCR Product Purification kit, Roche) and digested with BamHI. | Good bands. The 5’of the primers were designed with BamHI sites for easy circularization of the plasmid. The PCR product was cleaned (High Pure PCR Product Purification kit, Roche) and digested with BamHI. | ||
| + | |||
<pre> | <pre> | ||
- 22ul plasmid(~500ng) | - 22ul plasmid(~500ng) | ||
| Line 78: | Line 79: | ||
- 0.5ul BamHI (fermentas) | - 0.5ul BamHI (fermentas) | ||
</pre> | </pre> | ||
| + | |||
Digestion was cleaned (Roche kit) and a ligation was performed for circularization | Digestion was cleaned (Roche kit) and a ligation was performed for circularization | ||
| + | |||
<pre> | <pre> | ||
- 10ul plasmid(~40ng) | - 10ul plasmid(~40ng) | ||
| Line 85: | Line 88: | ||
- 30ul MQ | - 30ul MQ | ||
</pre> | </pre> | ||
| + | |||
20ul was used for a transformation of an E.coli top10 strain made competent using the CaCl2 method. Only a few colonies grew on the positive control transformation. Should be repeated with good competent cells. | 20ul was used for a transformation of an E.coli top10 strain made competent using the CaCl2 method. Only a few colonies grew on the positive control transformation. Should be repeated with good competent cells. | ||
Revision as of 23:29, 7 October 2010
We've started working on the wiki, been preparing plasmids and are settling in our office space. Note to self: should have brought coffee machine
Week 22, Arend Jan
The biobrick prefix and suffix are introduced into the pNZ8901 expression vector behind the spaS promoter using PCR and primers with 5’ overhangs. In this way, any biobrick with a RBS followed by a coding sequence can be brought to expression upon subtilin induction.
For optimization taq polymerase (fermentas) is used first.
PCR:
Component µl Final concentration Primer pNZ89bbs-for1 5 300nM Primer pNZ89bbs-rev1 5 300nM 10x Taq buffer(-MgCl2) 5 1x dNTP’s 2 200µM MgCl2 3 1.5mM Template 0.5 ~14ng Taq 0.5 2.5u MQ 29 - 94°C, 3’ - 94°C, 30’’ 30X - 50°C, 30’’ - 72°C, 1’30’’ - 72°C, 10’
No product. Likely the elongation step was too short for taq.
Repeat of previous PCR using the following cycling conditions.
- 94°C, 3’ - 94°C, 30’’ 30X - 50°C, 30’’ - 72°C, 3’ - 72°C, 10’
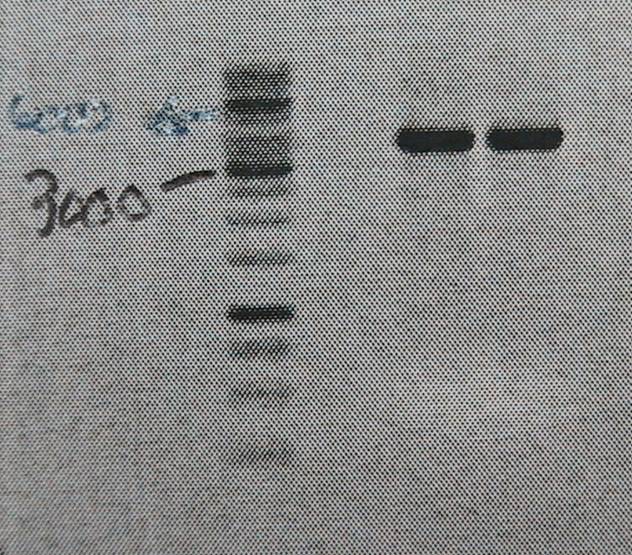
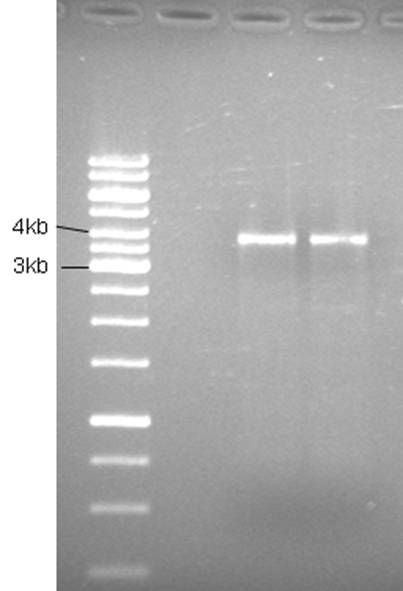
Product is ~4kb instead of 3,2. This has been observed before and plasmid is ok to use.
Repeated PCR with pfu polymerase (fermentas) to prevent point mutations in plasmid.
PCR:
Component µl Final concentration Primer pNZ89bbs-for1 5 300nM Primer pNZ89bbs-rev1 5 300nM 10x pfu buffer(-MgSO4) 5 1x dNTP’s 2 200µM MgCl2 3 1.5mM Template 0.5 ~14ng Pfu polymerase 1 2.5u MQ 28.5 - 94°C, 3’ - 94°C, 30’’ 30X - 50°C, 30’’ - 72°C, 1’30’’ - 72°C, 10’
Good bands. The 5’of the primers were designed with BamHI sites for easy circularization of the plasmid. The PCR product was cleaned (High Pure PCR Product Purification kit, Roche) and digested with BamHI.
- 22ul plasmid(~500ng) - 2.5ul buffer G - 0.5ul BamHI (fermentas)
Digestion was cleaned (Roche kit) and a ligation was performed for circularization
- 10ul plasmid(~40ng) - 5ul 10x T4 ligase buffer - 5ul T4 ligase (fermentas, LC) - 30ul MQ
20ul was used for a transformation of an E.coli top10 strain made competent using the CaCl2 method. Only a few colonies grew on the positive control transformation. Should be repeated with good competent cells.
 "
"