Team:ETHZ Basel/Modeling
From 2010.igem.org
(Difference between revisions)
(→Mathematical Modeling Overview) |
(→Mathematical Modeling Overview) |
||
| (17 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
= Mathematical Modeling Overview = | = Mathematical Modeling Overview = | ||
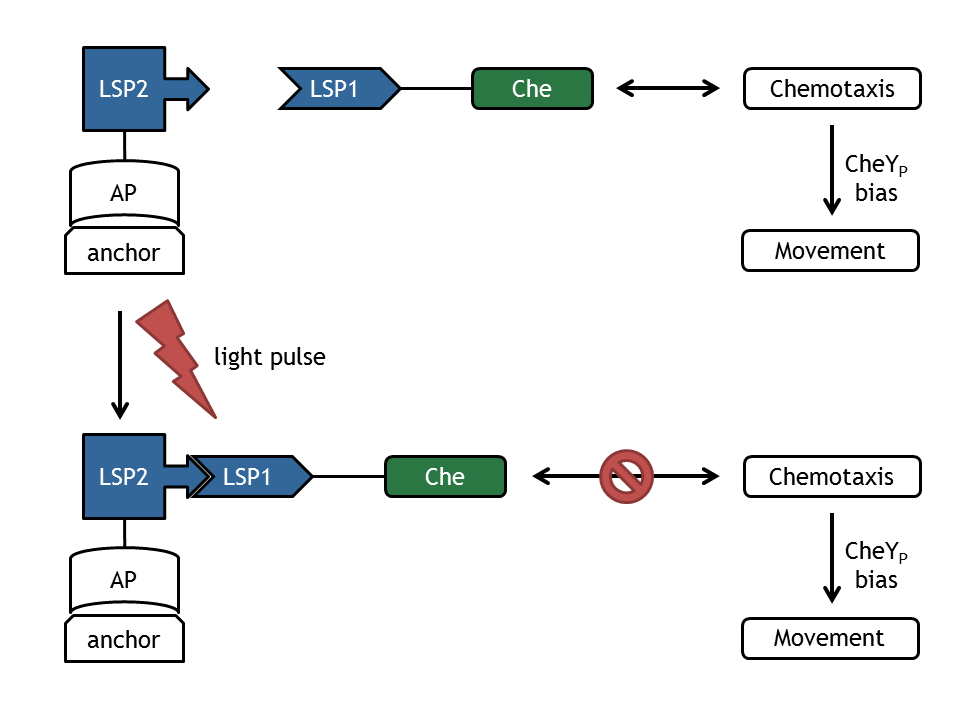
| - | [[Image:ETHZ_Basel_molecular_comb.png|thumb|400px|''' | + | [[Image:ETHZ_Basel_molecular_comb.png|thumb|400px|'''Figure 1: schematical overview of the modeled processes in E. lemming.''' LSP refers to light switch protein, AP to anchor protein, and Che to the attacked protein of the chemotaxis pathway.]] |
| - | A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created | + | A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created that covers the processes displayed in Figure 1. |
| - | + | In a first step, existing models for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrated the model with regard to available literature knowledge. | |
| + | * [[Team:ETHZ_Basel/Modeling/Light_Switch|'''Light Switch''']]: both implementation approaches have been modeled: | ||
| + | ** [[Team:ETHZ_Basel/Modeling/Light_Switch#Modeling_of_the_light_switch:_PhyB.2FPIF3|'''PhyB/PIF3''']]: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. | ||
| + | ** [[Team:ETHZ_Basel/Modeling/Light_Switch#Modeling_of_the_PhyB.2FPIF3_light_switch#Archeal_light_receptor|'''Archeal Light Receptor''']]: a deterministic molecular model based on the archeal light receptor. | ||
| + | * [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two deterministic molecular models of the chemotaxis pathway. | ||
| + | * [[Team:ETHZ_Basel/Modeling/Movement|'''Bacterial Movement''']]: a self developed stochastic model of ''E. coli'' movement on basis of the CheYp bias. | ||
| - | + | In a second part, we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to: | |
| - | + | * [[Team:ETHZ_Basel/Modeling/Combined#PhyB.2FPIF3_light_switch_-_Chemotaxis |'''PhyB/PIF3 light switch - Chemotaxis''']]: this model was used to reduce [[Team:ETHZ_Basel/Biology|wet laboratory experiments]] by identification molecular targets by [[Team:ETHZ_Basel/Modeling/Experimental_Design|experimental design]]. | |
| - | In a | + | * [[Team:ETHZ_Basel/Modeling/Combined#Archeal_light_receptor_-_Chemotaxis |'''Archeal light receptor - Chemotaxis''']]: this model was combined identically to the one above. |
| - | * [[Team:ETHZ_Basel/Modeling/ | + | * [[Team:ETHZ_Basel/Modeling/Combined#Chemotaxis_-_Movement |'''Chemotaxis - Movement''']]: complete model of E. lemming as a simulative test bench for the [[Team:ETHZ_Basel/InformationProcessing/Controller|controller]] design and as a brick of the comprehensive simulation of [[Team:ETHZ_Basel/InformationProcessing|information processing]]. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | * [[Team:ETHZ_Basel/Modeling/Combined# | + | |
| - | * [[Team:ETHZ_Basel/Modeling/Combined#Chemotaxis_-_Movement |''' Chemotaxis - Movement''']]: complete model of E. lemming | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Team:ETHZ_Basel/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Team:ETHZ_Basel/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 19:09, 27 October 2010
Mathematical Modeling Overview
A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created that covers the processes displayed in Figure 1.
In a first step, existing models for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrated the model with regard to available literature knowledge.
- Light Switch: both implementation approaches have been modeled:
- PhyB/PIF3: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3.
- Archeal Light Receptor: a deterministic molecular model based on the archeal light receptor.
- Chemotaxis Pathway: two deterministic molecular models of the chemotaxis pathway.
- Bacterial Movement: a self developed stochastic model of E. coli movement on basis of the CheYp bias.
In a second part, we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to:
- PhyB/PIF3 light switch - Chemotaxis: this model was used to reduce wet laboratory experiments by identification molecular targets by experimental design.
- Archeal light receptor - Chemotaxis: this model was combined identically to the one above.
- Chemotaxis - Movement: complete model of E. lemming as a simulative test bench for the controller design and as a brick of the comprehensive simulation of information processing.
 "
"