Team:ETHZ Basel/Biology
From 2010.igem.org
(→Biology & Wet Laboratory Overview) |
(→Biology & Wet Laboratory Overview) |
||
| Line 9: | Line 9: | ||
</html> | </html> | ||
| - | The | + | The core element of E. lemming is '''spatial localization''' of certain elements of the chemotactic network (Che proteins) and thus affecting the activity of their downstream partners. This anchoring is achieved with the help of '''light-sensitive proteins LSP's''' that dimerize upon a light signal. Dimerization results in spatial relocalization of the Che-protein and therefore altering the ratio between tumbling and directed flagellar movement. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Inside the cell, the chemotactic proteins CheA, CheY and CheZ tend to co-localize with methyl accepting chemotaxis protein MCPs at the membrane. But whereas CheA and CheZ nearly only localize at the MCPs, CheY is also present in significant concentrations in the cytoplasm making it's localization more straightforward [1]. | |
| - | + | For spatial localization of the Che-protein complex, three different '''anchor-proteins''' will be utilized: | |
| - | + | <br> 1. The '''tetracyclin repressor tetR''' anchoring the Che-protein to the DNA by binding to it's operator site tetO that has been inserted into a plasmid [2]. | |
| + | <br> 2. The '''triggor factor TrigA''' binding to the large ribosomal subunit [3]. | ||
| + | <br> 3. The '''prokaryotic actin homologue MreB''' which assembles into helical filaments underneath the cytoplasmic membrane [4]. | ||
| + | |||
| + | These anchors should enable to control the tumbling frequency by localizing a Che-protein and therefore interfering with its activity. | ||
| + | |||
| + | |||
| + | Remove all the rest? | ||
== generation of BioBricks == | == generation of BioBricks == | ||
Revision as of 10:17, 13 October 2010
Biology & Wet Laboratory Overview
The core element of E. lemming is spatial localization of certain elements of the chemotactic network (Che proteins) and thus affecting the activity of their downstream partners. This anchoring is achieved with the help of light-sensitive proteins LSP's that dimerize upon a light signal. Dimerization results in spatial relocalization of the Che-protein and therefore altering the ratio between tumbling and directed flagellar movement.
Inside the cell, the chemotactic proteins CheA, CheY and CheZ tend to co-localize with methyl accepting chemotaxis protein MCPs at the membrane. But whereas CheA and CheZ nearly only localize at the MCPs, CheY is also present in significant concentrations in the cytoplasm making it's localization more straightforward [1].
For spatial localization of the Che-protein complex, three different anchor-proteins will be utilized:
1. The tetracyclin repressor tetR anchoring the Che-protein to the DNA by binding to it's operator site tetO that has been inserted into a plasmid [2].
2. The triggor factor TrigA binding to the large ribosomal subunit [3].
3. The prokaryotic actin homologue MreB which assembles into helical filaments underneath the cytoplasmic membrane [4].
These anchors should enable to control the tumbling frequency by localizing a Che-protein and therefore interfering with its activity.
Remove all the rest?
generation of BioBricks
All utilized parts will be generated by PCR and subcloned into the storage vector pSEVA132 (Victor de Lorenzo's lab, KanR, BBR1 ori) allowing blue white screening. The working process for the generation of the subparts is as follows:
1. Ordering of primers (if template is available)
2. PCR
3. clean-up of PCR product
4. Ligation into storage vector
5. Transformation
6. Blue-white screening
7. Sequencing
Due to the presence of rare codons in the sequence of PhyB and Pif3, these two genes will be ordered from GenArt. However, as synthesizing takes several weeks, expression of the wild-type gene of these two proteins will be tested and if satisfying proceeded with these constructs.
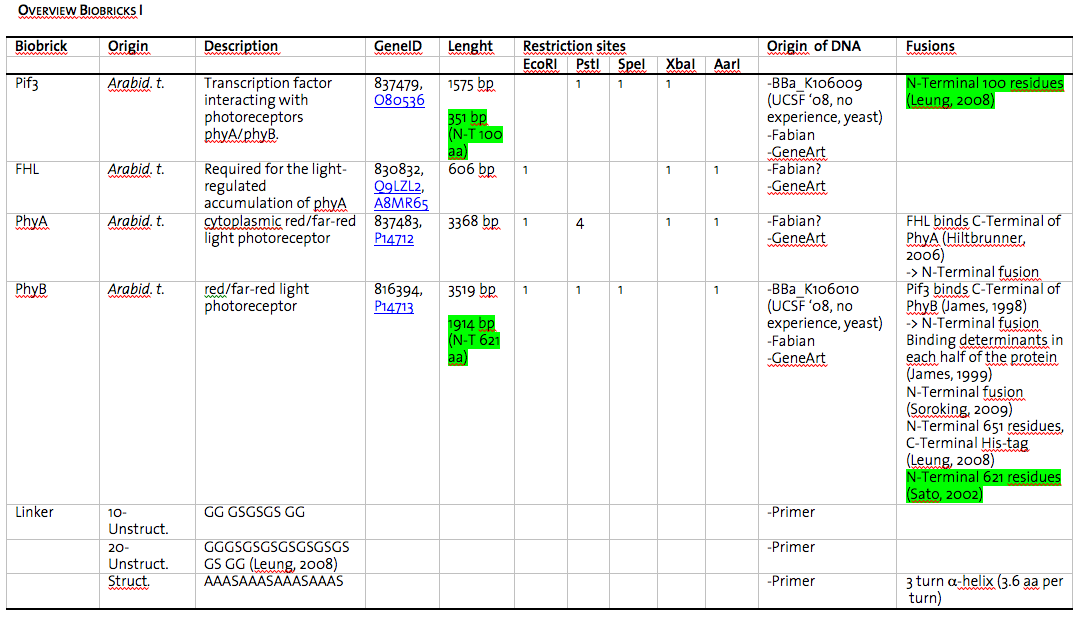
In this picture all the details (origin of DNA, length of gene, restriction sites, sources) about the BioBricks generated are displayed.
generation of fusion proteins
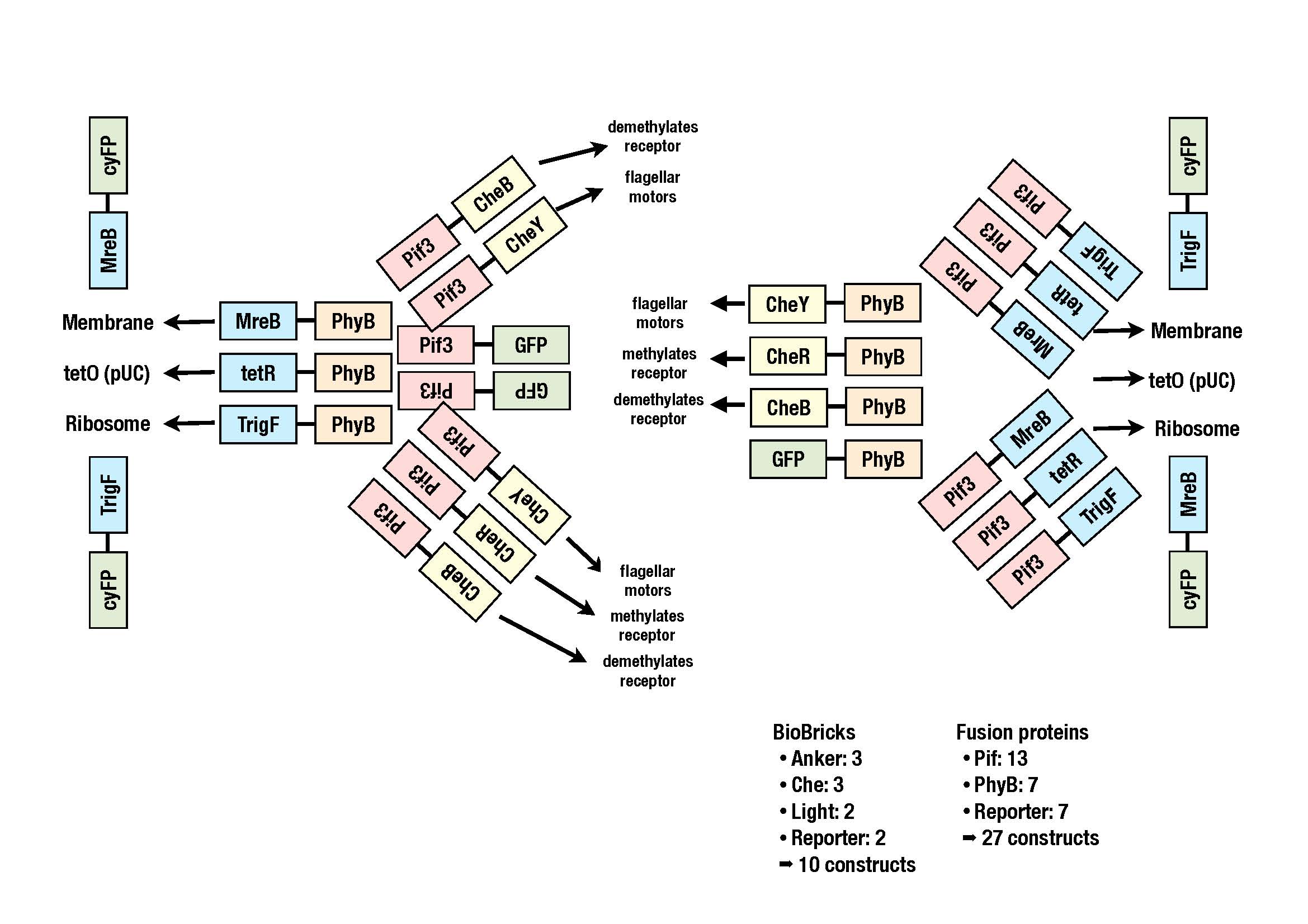
The image shows all the constructs we plan to clone.Once the BioBricks in the storage vectors are available, the fusion proteins in the acceoptor vector will be generated according to the cloning strategy BBF RFC 28.
Functionality assays
The constructs will have to be tested for the following properties:
- Che protein fusion: Influencing of chemotactic network
- Localizer fusion: Spatial localization
- PhyB-Pif system: Activation by light
Chemotactic Functionality
Is the Che fusion protein is still functional?
Possible assays to investigate the functionality:
Localization
Is the localizer spatially separating PIF3 or PhyB to a certain area within the cell?
Fluorescence microscopy will be utilized to answer this question. Therefore, fusions of the anchor to fluorescent proteins (cyfp, gfp) have to be generated.
 "
"