Team:ETHZ Basel
From 2010.igem.org
| Line 27: | Line 27: | ||
!align="center"|[[Team:ETHZ_Basel/ImagingPipeline|Imaging Pipeline]] | !align="center"|[[Team:ETHZ_Basel/ImagingPipeline|Imaging Pipeline]] | ||
!align="center"|[[Team:ETHZ_Basel/Notebook|Notebook]] | !align="center"|[[Team:ETHZ_Basel/Notebook|Notebook]] | ||
| - | !align="center"|[[Team:ETHZ_Basel/ | + | !align="center"|[[Team:ETHZ_Basel/Literature|Literature]] |
|} | |} | ||
Revision as of 14:11, 24 July 2010
E. lemming
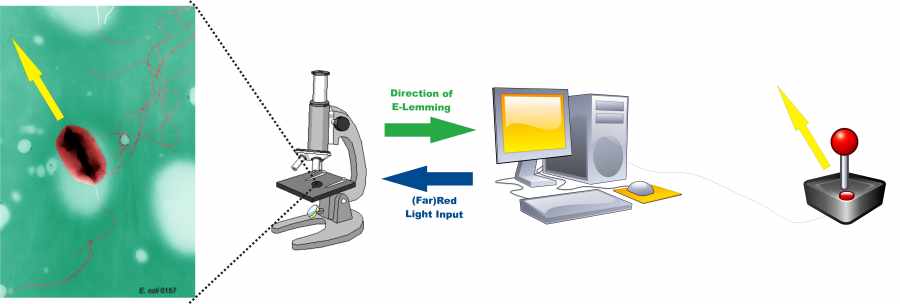
ETHZ Basel project goal is to control E. coli movements (chemotaxis) by means of light. In fact, we will change the chemotaxis pathway either by substituting the receptor with a light-sensitive one or by interfering with the kinase-phosphatase process with proteins whose binding and unbinding can be stimulated by pulses of light. In both ways, E. coli tumbling is induced or removed just by pressing a light switch and, as a consequence, a bacterium can be "driven" to a precise, pre-fixed point. Tumbling / directed flagellar movement rates are supervised by digital controller combined with image processing algorithms. This system enables to control single E. coli cells to move like mindless "Lemmings" in the direction they are forced to go.
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Imaging Pipeline | Notebook | Literature |
|---|
 "
"