Team:Debrecen-Hungary/minimals
From 2010.igem.org
(→The Tetracycline On/Off Gene Expression System) |
(→The Tetracycline On/Off Gene Expression System) |
||
| Line 101: | Line 101: | ||
The Tet-On system (c) works in the opposite way so the rtTA protein is capable of binding the operator only when bound by doxycycline. Thus the introduction of doxycyline to the system initiates the transcription of the genetic product. The Tet-On system is sometimes preferred for the faster responsiveness. | The Tet-On system (c) works in the opposite way so the rtTA protein is capable of binding the operator only when bound by doxycycline. Thus the introduction of doxycyline to the system initiates the transcription of the genetic product. The Tet-On system is sometimes preferred for the faster responsiveness. | ||
| - | [[Image:Tet system.jpg|800px|thumb|center]] | + | [[Image:Tet system.jpg|800px|thumb|center|]] |
''The alternative version of the original Tet-On and Tet-Off Systems called Tet-Advanced have been optimized for improved expression in mammalian cells by utilizing human codon preferences and removing cryptic splice sites from the mRNA sequence. These enhancements lead to higher and more stable expression levels, minimize off-target effects and reduce toxicity.'' | ''The alternative version of the original Tet-On and Tet-Off Systems called Tet-Advanced have been optimized for improved expression in mammalian cells by utilizing human codon preferences and removing cryptic splice sites from the mRNA sequence. These enhancements lead to higher and more stable expression levels, minimize off-target effects and reduce toxicity.'' | ||
Revision as of 09:35, 26 October 2010
|
Welcome To The MinimalsFrom year to year (and from one to jamboree to another) the world of synthetic biology exponentially expands. Some iGEMers may find niches of complex biological systems and use special model organisms or laboratory tools. Our philosophy is that a good project is one that can be kept simple and short.
ContentsScientific Background Cellular signaling - Nuclear Receptors - Structure of NRs - Apoptosis - The Tetracycline On/Off Gene Expression System Model Organisms Drosophila Melanogaster - Caenorhabditis elegans - Homo sapiens In The Laboratory (Techniques And Reagents) Cotransfection Assay, Two Hybrid Assay - Luciferase - Cos-1 cells - Dose response curve Scientific BackgroundCellular signalingCells have an innate ability to “listen” and correctly react to their local or even distant environment. Through time it has been observed that a complex systems of communication governs essential cellular activates and coordinates cell actions.[1] Today, it is well known that processes such as development, growth, tissue repair or death, metabolic shifts and immunity are all governed, at the molecular level, by signaling. By understanding cell signaling, diseases may be treated effectively and, theoretically, artificial tissues may be created. Cells sense information from their local surroundings through a class of proteins known as receptors. Chemicals that activate (or inhibit) receptors are often named hormones, growth factors, cytokines or even neurotransmitters yet their proper term is receptor ligands. Water soluble ligands have cell membrane penetration and thus mostly interact with trans-membranous receptors, whereas ligands with high lipid solubility easily penetrate the cell membrane
Nuclear Receptors
Structure of NRsLigand binding domain (LBD) is a well conserved domain amongst various nuclear receptors whose structure usually referred to as an alpha helical sandwich fold. The LBD shows some diversity among nuclear receptors as it is a site for receptor-specific events. It possesses transactivation ability and contains a ligand-binding pocket as well as the main interaction surfaces for other proteins [8]. The DNA-binding domain (DBD) contains two Zinc-finger motifs and is linked to the LBD by a highly flexible hinge region. This segment of the nuclear receptors holds the ability to recognize and bind to preferred or specific DNA-motifs, the response elements. A partially unexpected and amazing feature of the nuclear receptors is that the above mentioned domains can be swapped. It was Vincent Giguere and his colleagues, who first devised a technique called cotransfection assay and developed it to the domain-swap technique.
ApoptosisApoptosis, the process of programmed cell death (PCD), is an elaborate cellular homeostasis mechanism that ensures correct development and function of multicellular organisms. Biochemical events lead to characteristic changes (morphology) and the death of cells. It has a crucial role in maintaining tissue homeostasis as provides the appropriate rate of cellular turnover. Potentially dangerous, poorly differentiated or excess cells are removed by apoptosis without local inflammation from leakage of cell contents. Dying by apoptosis in many cases has primary importance: lens cells, which lost their nucleus, need the vision, the corneocytes without nucleus and organelles provide mechanical protection on the outer surface of the skin. Apoptosis might occur in pathological conditions, mainly in secondary, as a part of the defensive mechanism, helping to eliminate the viral infected or malignant cells. The abnormality of the central molecules and genes of apoptosis may play a role in the development of certain diseases: degenerative disease, autoimmune disease, tumor formation and malformations. Many pathways and signals lead to apoptosis, but there is only one mechanism that actually causes the death of a cell. After a cell receives stimulus, it undergoes organized degradation of cellular organelles by activated proteolytic caspases. A cell undergoing apoptosis shows characteristic morphology.
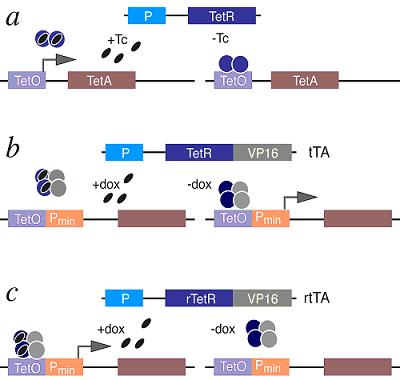
The Tetracycline On/Off Gene Expression SystemThe most commonly used inducible expression systems for research of eukaryote cell biology are named Tet-Off and Tet-On called togheter the Tetracyclin-controlled transcriptional activation. Gene expression is controlled as a result of binding of the Tet-Off (tetracycline transactivator) or Tet-On (reverse tetracycline transactivator) protein to tetracycline response elements (TREs) located within an inducible promoter such as minimal Cytomegalovirus (CMV) promoter. Each system consists of 2 components (a) that have been optimized for use in mammalian cells: /1/ A regulator vector which expresses one of the tetracycline-controlled transactivators. /2/ A response vector containing TRE within the promoter that controls expression of the gene of interest. The Tet-Off system (b) makes use of the tetracycline transactivator (tTA) protein created by fusing TetR (tetracycline repressor) found in E. coli with another protein, VP16 produced by the Herpes Simplex Virus. The tTA binds the DNA at a tet-o operator. Once bound to the tet-o the tTA will activate a promoter coupled to the tet-o operator, activating the transcription of the gene of interest. Tetracycline derivatives bind tTA and render it incapable of binding to TRE thereby preventing transactivation of target genes. The TetR can prevent alone the expression of the nearby gene and the presence of tetracylin can force the promoter effect before the gene of interest. The Tet-On system (c) works in the opposite way so the rtTA protein is capable of binding the operator only when bound by doxycycline. Thus the introduction of doxycyline to the system initiates the transcription of the genetic product. The Tet-On system is sometimes preferred for the faster responsiveness. The alternative version of the original Tet-On and Tet-Off Systems called Tet-Advanced have been optimized for improved expression in mammalian cells by utilizing human codon preferences and removing cryptic splice sites from the mRNA sequence. These enhancements lead to higher and more stable expression levels, minimize off-target effects and reduce toxicity. Model OrganismsDrosophila Melanogaster
Drosophila Melanogaster, also known as the common fruit fly, is one of the most frequently used model organisms in biological sciences, including studies in genetics, physiology, microbial pathogenesis and life history evolution.[9] The ecdysone receptor is a nuclear receptor found in D.Melanogaster, where it controls development and contributes to other processes such as reproduction. Its ligands are ecdysteroid which are secreted by the organism’s prothoracic gland.
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode (roundworm), about 1 mm in length,[10] which lives in temperate soil environments C. elegans is intensively studied as a model organism in biology for a variety of reasons. The developmental fate of every single somatic cell (959 in the adult hermaphrodite; 1031 in the adult male) has been mapped out.[11] [12] The C.elegans genome harbors 284 nuclear receptors [10] (a striking figure), which have been shown to control traits such as sex determination, larva development, life span, neuronal growth and identity and much more. As far as nuclear receptors go, they are a gold mine.
Homo SapiensHomo sapiens are the only living species in the Homo genus of bipedal primates in the great ape family. Nuclear receptors number up to 47 in humans, yet only few have been well characterized. They constitute the focus of medicinal reproductive technologies, hormonal medicine (endocrinology), immunology, drug interaction and much more.
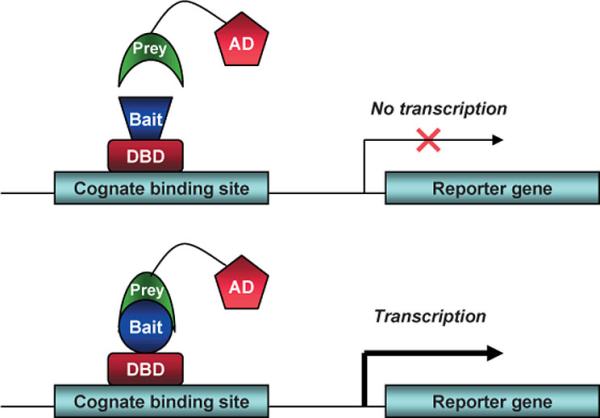
In The Laboratory (Techniques And Reagents)Cotransfection Assay, Two Hybrid AssayCotransfection assay requires two types plasmids to be cotransfected into a cell; an expression vector, coding a functional NR and and a reporter-plasmid, harboring an inducible promoter that regulates a reporter-gene the expression level of which can easily be detected and quantified. By this method the NR-activity and ligand-potency can be studied. As a result of the unique domain-structure of NRs, the domain-swap technique was developed, using chimeric proteins; the DBD of a known receptor is fused to any other receptor. This made ligand screening possible. The two-hybrid assay is a technique in molecular biology which can be used to investigate protein-protein interactions[13] [14]. The interaction of a DNA-binding bait-protein and the prey-protein with a ligand binding and activating ability is shown by the activated reporter.
- the response elements by changing the sequence that is recognized by the DBD - potential dimer partners and co-regulators by creating specific bait-prey sets - potential ligands and activating factors
This image shows the basic concept of the two-hybrid system: the DNA-binding bait-protein (Gal-fused, in our case), and the activator-domain fused to the prey-protein (mostly VP-fusion, in our case) and the reporter gene with the specific binding site. Yeast Gal4 is a common DBD used for this techniques purpose. Commly used reporter genes include the product of the LacZ gene (Beta galactosidase) and Luciferase. Luciferase
Luciferase is an enzyme class able to produce bioluminescence by oxidizing the substrate luciferin. "Firefly luciferase" as a laboratory reagent usually refers to P. pyralis luciferase. Its emission can be measured photometrically and hence used to deduce the protein enzyme concentration through standardized methods.
Cos-1 cells
Cos-1 cells (acronym for CV-1 simian origin, SV-40 viral positive) is cell line derived from the African green monkey kidney cells. It is also often used to transfect cells in tissue culture conditions to produce recombinant proteins for molecular biology, biochemistry, and cell biology experiments. Two forms of COS cell lines commonly used are COS-1 and COS-7. The cell line was obtained by immortalizing the original CV-1 cells with SV-40 virus genome. This allows the production of large T antigen but has a defect in genomic replication.[15] Dose response curveDose response curve depicts a change in a measured effect on an organism caused by differing levels of exposure to a chemical in standardized measuring conditions. It may apply to either individuals or to populations. The curve is usually displayed in a simple X-Y graph (X being logarithm of dose, Y for effect). The half maximal effective concentration (EC50), a common feature of drug potency, is the chemical’s concentration which induces a response halfway between the baseline and maximum.[16]
References
1. ^ Witzany, Guenther (2010). Biocommunication and Natural Genome Editing. Springer. ISBN 9789048133185. 
|
 "
"
 Team
Team