|
|
| (48 intermediate revisions not shown) |
| Line 100: |
Line 100: |
| | /* End IE5-Mac hack */ | | /* End IE5-Mac hack */ |
| | #tabs9 a:hover span { | | #tabs9 a:hover span { |
| | + | background-color: #ff3366; |
| | color:#FFFFFF; | | color:#FFFFFF; |
| | } | | } |
| Line 236: |
Line 237: |
| | </div> | | </div> |
| | </body> | | </body> |
| | + | </html> |
| | <!--- 2nd tab End---> | | <!--- 2nd tab End---> |
| - | <!--- 3rd tab--->
| |
| - | <div id="search-controls">
| |
| - | <form action="/Special:Search" id="searchform">
| |
| - | <input id="searchInput" name="search" type="text" title="Search 2010.igem.org [f]" accesskey="f" value="" />
| |
| - | <input type='submit' name="go" class="searchButton" id="searchGoButton" value="Go" title="Go to a page with this exact name if exists" />
| |
| - | <input type='submit' name="fulltext" class="searchButton" id="mw-searchButton" value="Search" title="Search the pages for this text" />
| |
| - | </form>
| |
| - | </div> <!-- close search controls -->
| |
| - | <p>
| |
| - | <style type="text/css">
| |
| - | <!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
| |
| - | <html xmlns="http://www.w3.org/1999/xhtml">
| |
| - | <head>
| |
| - | <meta http-equiv="Content-Type" content="text/html; charset=iso-8859-1" />
| |
| - | <title>Free CSS Navigation Menu Designs 2 at exploding-boy.com</title>
| |
| - | <style type="text/css">
| |
| - | <!--
| |
| - | body {
| |
| - | margin:0;
| |
| - | padding:0;
| |
| - | font: bold 11px/1.5em Verdana;
| |
| - | }
| |
| - |
| |
| - | h2 {
| |
| - | font: bold 14px Verdana, Arial, Helvetica, sans-serif;
| |
| - | color: #000;
| |
| - | margin: 0px;
| |
| - | padding: 0px 0px 0px 15px;
| |
| - | }
| |
| - |
| |
| - | img {
| |
| - | border: none;
| |
| - | }
| |
| - |
| |
| - | /*- Menu Tabs 8--------------------------- */
| |
| - |
| |
| - | #tabs8 {
| |
| - | float:left;
| |
| - | width:100%;
| |
| - | font-size:100%;
| |
| - | line-height:normal;
| |
| - | background:#007500;
| |
| - | margin-top:0px;
| |
| - | }
| |
| - | #tabs8 ul {
| |
| - | margin:0;
| |
| - | padding:2px 10px 0 105px;
| |
| - | list-style:none;
| |
| - | }
| |
| - | #tabs8 li {
| |
| - | float:left;
| |
| - | display:inline;
| |
| - | margin:0;
| |
| - | padding:0;
| |
| - | margin-left:40px;
| |
| - | }
| |
| - | #tabs8 a {
| |
| - | float:left;
| |
| - | background:url("tableft10.gif") no-repeat left top;
| |
| - | margin:0;
| |
| - | padding:0 0 0 2px;
| |
| - | text-decoration:none;
| |
| - | }
| |
| - | #tabs8 a span {
| |
| - | float:left;
| |
| - | display:block;
| |
| - | background:url("tabright10.gif") no-repeat right top;
| |
| - | padding:5px 15px 4px 6px;
| |
| - | color:#FFF;
| |
| - | }
| |
| - | /* Commented Backslash Hack hides rule from IE5-Mac \*/
| |
| - | #tabs8 a span {float:none;}
| |
| - | /* End IE5-Mac hack */
| |
| - | #tabs8 a:hover span {
| |
| - | color:#FFFFFF;
| |
| - | }
| |
| - | #tabs8 a:hover {
| |
| - | background-position:0% -42px;
| |
| - | }
| |
| - | #tabs8 a:hover span {
| |
| - | background-position:100% -42px;
| |
| - | }
| |
| - |
| |
| - | #tabs8 #current a {
| |
| - | background-position:0% -42px;
| |
| - | }
| |
| - | #tabs8 #current a span {
| |
| - | background-position:100% -42px;
| |
| - | }
| |
| - | -->
| |
| - | </style>
| |
| - | </head>
| |
| - |
| |
| - | <body>
| |
| - | <div id="tabs8">
| |
| - | <ul>
| |
| - | <!-- CSS Tabs -->
| |
| - | <li><a href="https://2010.igem.org/Team:Chiba/System_2/Testing_components"><span>Testing</span></a></li>
| |
| - | <li><a href="https://2010.igem.org/Team:Chiba/System_2/Construction_Process"><span>Construction</span></a></li>
| |
| - | <li><a href="https://2010.igem.org/Team:Chiba/System_2/Evaluation_subsystem"><span>Evaluation</span></a></li>
| |
| - | <li><a href="https://2010.igem.org/Team:Chiba/System_2/Result"><span>Result</span></a></li>
| |
| - | <li><a href="https://2010.igem.org/Team:Chiba/System_2/Remarks"><span>Remarks</span></a></li>
| |
| - | </ul>
| |
| - | </div>
| |
| - | </body>
| |
| - | <!--- 3rd tab End --->
| |
| | <!--- Background Contents End ---> | | <!--- Background Contents End ---> |
| | <!-- ------------------------------------------------------------------------------- | | <!-- ------------------------------------------------------------------------------- |
| Line 349: |
Line 245: |
| | ------------------------------------------------------------------------------- | | ------------------------------------------------------------------------------- |
| | ----------------------------------------------------------------------------------- --> | | ----------------------------------------------------------------------------------- --> |
| - | <br><br><br><br><br><br>
| |
| - | <font size=6>Version 2 :</font>
| |
| | <!-------- Main --------> | | <!-------- Main --------> |
| - | <html>
| + | =Version 2= |
| - | <table border="0" cellpadding="30" cellspacing="0">
| + | |
| - | <tr>
| + | |
| - | <td width="965px"><font size="4" face="verdana">Overall Circuit</font><br>
| + | |
| - | <hr width="500" size="1" align="left"><br>
| + | |
| - | <center>
| + | |
| - | <font size=2 face=verdana>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2010/1/1a/Chiba_Sys2.jpg"><br>
| + | |
| - | </center>
| + | |
| - | Our second circiut of double click system is called <b>Recognize circuit</b>.<br>
| + | |
| - | It consists of <b>recognizing input</b> system and <b>memorizing input</b> system.
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | <table border="0" cellpadding="30" cellspacing="0">
| + | |
| - | <tr>
| + | |
| - | <td width="965px"><font size="4" face="verdana">Control of circuit</font><br>
| + | |
| - | <hr width="500" size="1" align="left"><br>
| + | |
| - | <center>
| + | |
| - | <font size=2 face=verdana>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2010/9/99/Chiba_Sys2_Input.jpg"><br>
| + | |
| - | </center>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | <table border="0" cellpadding="30" cellspacing="0" margin-top="-50">
| + | |
| - | <tr>
| + | |
| - | <td width="400px"><font size=2 face=verdana>
| + | |
| - | We use AHL for input and regard injection of AHL as clicking. LuxR is generated constitutive. Injecting AHL to Double Click Bacteria brings dimer of AHL and luxR. We take lux promoter repressed by the dimer. As the dimer binds Plux, transcription of proteins under the Plux is repressed.<br>
| + | |
| - | So in our project, <br>
| + | |
| - | <b><font color="red">CLICK controls transcription of overall DNA sequence.</font></b>
| + | |
| - | </td>
| + | |
| - | <td width="400px"><center>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2010/e/e9/Chiba_LuxR.jpg"></center>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| | | | |
| - | <table border="0" cellpadding="30" cellspacing="0">
| + | ==Overview== |
| - | <tr>
| + | {|- |
| - | <td width="965px">
| + | |Our second circiut of double click system is called <b>Recognize circuit</b>.<br> |
| - | <font size="4" face="verdana">Recognizing Input System</font><br>
| + | It consists of <b>recognizing</b> system and <b>memorizing</b> system.<br> |
| - | <hr width="500" size="1" align="left"><br>
| + | Recognizing system can recognize the existence of input.<br> |
| - | <center>
| + | And Memorizing system can memorize the existence of input in the past for a while.<br> |
| - | <img src="
| + | When the two system work together, bacteria is going to shine.<br> |
| - | https://static.igem.org/mediawiki/2010/c/c8/Chiba_Sys2_Rec.jpg"><br>
| + | We hope this circuit works as double click system. |
| - | </center> | + | |[[Image:Chiba_cI.jpg]] |
| - | </td> | + | |} |
| - | </tr> | + | |
| - | </table> | + | |
| - | <table border="0" cellpadding="30" cellspacing="0" margin-top="-50"> | + | |
| - | <tr> | + | |
| - | <td width="400px"><font size=2 face=verdana>
| + | |
| - | In recognizing input system, there is a cI operator above gfp DNA sequence. If cI binds cI operator, transcription of gfp is stopped. You can see DNA sequence of cI, which a kind of repressor, is under the Plux. On initial state(there isn't any click), cI is generated normally. But AHL come into Double Click Bacteria, generation of cI is stopped. Since repression of Plux starts working by the dimer which is a combination of AHL and LuxR. And then, cI is going to disappear, because all kinds of proteins are bound to be decomposed. If there isn't cI anymore, bacteria will have chance to shine.<br>
| + | |
| - | We can say<br> | + | |
| - | <b><font color="red">cI recognizes the existence of input</font></b> in this DNA sequence.</td>
| + | |
| - | <td width="400px"><center>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2010/9/90/Chiba_cI.jpg"></center>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| | | | |
| | + | We use AHL for input and regard injection of AHL as clicking. In our project, CLICK controls transcription of overall DNA sequence. <br> |
| | + | In recognizing input system, there is a cI operator above gfp DNA sequence. cI recognizes the existence of input in this DNA sequence.<br> |
| | + | In memorizing system, there is an AND Gate with T7ptag and supD. In this case, the AND Gate remembers that there was a click.<br> |
| | + | There is a hybrid promoter(cI/T7) above gfp DNA sequence. It is regulated by T7 and cI which work as activator and repressor, respectively. On this promoter, repression is stronger than activation. And also, it is low unregulated activation. Working of this promoter depends on recognizing and memorizing system.<br> |
| | + | [[Team:Chiba/System_2/Overall|details...]] |
| | + | <br><br><br> |
| | | | |
| - | <table border="0" cellpadding="30" cellspacing="0">
| + | ==Circuit Construction== |
| - | <tr>
| + | We've designed DNA sequence like following Figure. <br> |
| - | <td width="965px">
| + | |
| - | <font size="4" face="verdana">Memorizing Input System</font><br>
| + | |
| - | <hr width="500" size="1" align="left"><br>
| + | |
| | <center> | | <center> |
| - | <img src="https://static.igem.org/mediawiki/2010/2/26/Chiba_Sys2_AND.jpg"><br>
| + | [[Image:Chiba_Sys2.jpg]]</center> |
| - | </center> | + | Based on this sequence, we've also designed vector.<br> |
| - | </td> | + | <center> |
| - | </tr> | + | [[Image:Chiba_ver2.jpg]]</center> |
| - | </table> | + | Prom-luxR&PT7/cI-GFP vector is constructed collaboration with Double Click System Version 1.<br><br><br> |
| - | <table border="0" cellpadding="30" cellspacing="0" margin-top="-50">
| + | |
| - | <tr>
| + | |
| - | <td width="400px"><font size=2 face=verdana>
| + | |
| - | In memorizing system, there is an AND Gate with T7ptag and supD. As you know, T7ptag becomes T7 polymerase in case of existing supD. T7 polymerase is translated to T7 protein which works as activator of T7 promoter. There is a T7 promoter above gfp DNA sequence. If T7 have been generated once, the T7 promoter starts translating. And then we can see bacteria begin to shine.<br><br>
| + | |
| | | | |
| | + | ==Result== |
| | | | |
| | + | ===Testing=== |
| | + | In this system, lux promoter inverter is sensor of input. |
| | + | We've researched Plux inverter and got some results.<br> |
| | + | We need lux promoter repressed by AHL-luxR dimer. But Plux inverter of biobrick didn't work. So we've tried to charaterize it and make Plux inverter.<br> |
| | + | LuxR is AHL-dependent activator. LuxR-AHL complex binds lux box, 20-bp sequence centered at position -42.5 from starting site and activates transcription. However lux box is inserted between -35 and -10,LuxR functions as AHL-inverter (Plux inv). Plux inv is resistered in Biobrick number R0061. We've prepared Plux inv-GFP and characterized about it. [[Team:Chiba/System_2/Result|details...]] |
| | | | |
| - | On initial state, bacteria don't have supD. Because, lacI represses the lac promoter above supD DNA sequence.So there isn't any T7 polymerase and gfp cannot be genernated, in spite of being T7ptag in bacteria.<br><br>
| + | ===Evaluation=== |
| | + | <br> |
| | + | ===Remark&Notes=== |
| | + | <br> |
| | | | |
| - | | + | ==Conclusion== |
| - | | + | It does not completely repress transcription, Plasmids, strains ,culture conditions,and detection method LuxR suppression can not be confirmed(Cox et al,2007).<br> |
| - | As written above, when AHL is entered into bacteria, transcription of proteins under the Plux is repressed. If there is click, transcription of T7ptag and lacI are stopped simultaneously. But T7ptag are decomposed earlier than lacI because of difference in velocity of decomposition. T7ptag which is a kind of mRNA is decomposed so fast and lacI which is just a kind of protein is decomposed late. SupD under the Plac is generated after most of lacI are decomposed. So, during the clicking, there isn't any T7ptag as far as there is supD. <br><br>
| + | [[Team:Chiba/System_2/Result|details...]] |
| - | | + | <br><br><br> |
| - | | + | |
| - | | + | |
| - | After washing AHL(means after 1st click), supD exist. SupD is a kind of tRNA and stable which means having long half-life. If there isn't input anymore, Plux starts activating and T7ptag is generated. As result of reacting T7ptag and supD, the AND Gate is going to be available. In this case, <b><font color="red">the AND Gate remembers that there was a click.</font></b><br><br>
| + | |
| - | | + | |
| - | | + | |
| - | Moreover, if there is 2nd click, T7ptag is going to be decomposed again. And then, the reaction of And Gate is stopped, because there isn't T7ptag anymore. So, T7 which is activator of T7 promoter is also going to be decomposed. It is time-limit of Double-Click and explain it in next paragraph. </td>
| + | |
| - | <td width="400px"><center>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2010/6/6e/Chiba_third.jpg"></center>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <table border="0" cellpadding="30" cellspacing="0">
| + | |
| - | <tr>
| + | |
| - | <td width="965px">
| + | |
| - | <font size="4" face="verdana">Co-working of two systems </font><br>
| + | |
| - | <hr width="500" size="1" align="left"><br>
| + | |
| - | <center>
| + | |
| - | <img src="
| + | |
| - | https://static.igem.org/mediawiki/2010/b/b3/Chiba_Sys2_Whole.jpg"><br>
| + | |
| - | </center>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | <table border="0" cellpadding="30" cellspacing="0" margin-top="-50">
| + | |
| - | <tr>
| + | |
| - | <td width="900px"><font size=2 face=verdana>
| + | |
| - | Actually, there is a hybrid promoter(cI/T7) above gfp DNA sequence. It is regulated by T7 and cI which work as activator and repressor, respectively.
| + | |
| - | On this promoter, repression is stronger than activation. And also, it is low unregulated activation. <font color="red"><b>When 1st input</b></font> is come in, cI recognizes the input and going to be decomposed.
| + | |
| - | But there isn't T7 yet, <font color="red"><b>gfp cannot be generated</b></font>, despite there isn't repressor. <font color="red"><b>After 1st input,</b></font> AND gate remembers there was a click, and generation of T7 is stared. But, <font color="red"><b>gfp cannot be generated</b></font>, because there is cI already. <font color="red"><b>When 2nd input</b></font> is come, cI also recognizes the input and going to be decomposed. If the 2nd input comes within time that the AND is working, T7 exists in bacteria and decomposition of cI is finished, <font color="red"><b>gfp is going to be generated.</b></font>As a result, bacteria shines with gfp and Double Click is completed.</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | </html>
| + | |
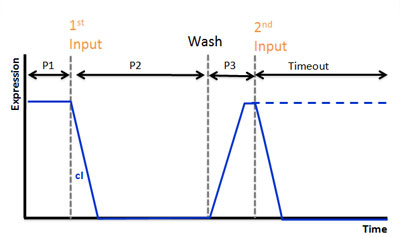
We use AHL for input and regard injection of AHL as clicking. In our project, CLICK controls transcription of overall DNA sequence.
In recognizing input system, there is a cI operator above gfp DNA sequence. cI recognizes the existence of input in this DNA sequence.
In memorizing system, there is an AND Gate with T7ptag and supD. In this case, the AND Gate remembers that there was a click.
There is a hybrid promoter(cI/T7) above gfp DNA sequence. It is regulated by T7 and cI which work as activator and repressor, respectively. On this promoter, repression is stronger than activation. And also, it is low unregulated activation. Working of this promoter depends on recognizing and memorizing system.
details...
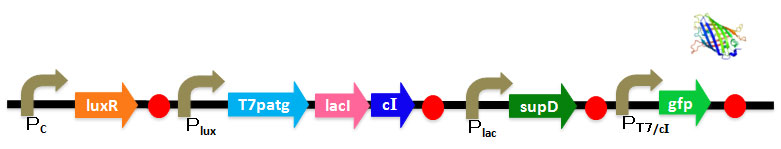
We've designed DNA sequence like following Figure.
Based on this sequence, we've also designed vector.
Prom-luxR&PT7/cI-GFP vector is constructed collaboration with Double Click System Version 1.
In this system, lux promoter inverter is sensor of input.
We've researched Plux inverter and got some results.
We need lux promoter repressed by AHL-luxR dimer. But Plux inverter of biobrick didn't work. So we've tried to charaterize it and make Plux inverter.
LuxR is AHL-dependent activator. LuxR-AHL complex binds lux box, 20-bp sequence centered at position -42.5 from starting site and activates transcription. However lux box is inserted between -35 and -10,LuxR functions as AHL-inverter (Plux inv). Plux inv is resistered in Biobrick number R0061. We've prepared Plux inv-GFP and characterized about it. details...
It does not completely repress transcription, Plasmids, strains ,culture conditions,and detection method LuxR suppression can not be confirmed(Cox et al,2007).
details...

 "
"