Team:Cambridge/Quiescence
From 2010.igem.org
Peteremmrich (Talk | contribs) (→Quiescence) |
Peteremmrich (Talk | contribs) (→The aptamer-controlled-ribozyme approach) |
||
| Line 16: | Line 16: | ||
A new approach to making a quiescence switch would be to control the functionality of the folded RNA instead of its transcription, which proved to be problematic. Ideally the bacteria would grow in the lab or a vat in the presence of a ligand but would stop growing without dying, as soon as the ligand is removed. Thus if the engineered bacteria escaped into the wild they would stop growing very quickly as the ligand is diluted. | A new approach to making a quiescence switch would be to control the functionality of the folded RNA instead of its transcription, which proved to be problematic. Ideally the bacteria would grow in the lab or a vat in the presence of a ligand but would stop growing without dying, as soon as the ligand is removed. Thus if the engineered bacteria escaped into the wild they would stop growing very quickly as the ligand is diluted. | ||
| - | We hope to achieve this by rationally designing a gene for an RNA that folds into a non-functional shape. The structure will include a hammerhead ribozyme domain (see | + | We hope to achieve this by rationally designing a gene for an RNA that folds into a non-functional shape. The structure will include a hammerhead ribozyme domain (see [http://sage.ucsc.edu/scottlab/reprints/2006_Scott_Cell_pdf/2006_Scott_Cell.pdf Martick and Scott 2006]) that will hopefully cleave itself in vivo to release the functional Rcd RNA. In order to control this cleavage, the final construct would include an aptamer, which is an RNA structure able to bind a specific compound. This would create an allosteric ribozyme. |
The project would involve synthesising different constructs (which are all mercifully short) and testing their functionality and sensitivity. The result would be a fairly short, but quite versatile BioBrick. | The project would involve synthesising different constructs (which are all mercifully short) and testing their functionality and sensitivity. The result would be a fairly short, but quite versatile BioBrick. | ||
Revision as of 14:17, 28 July 2010

Quiescence
This Project seeks to design a device that can be used to switch cells into a non-growing, but metabolically active state. A system like this would be highly desirable, as it could be used to increase the efficiency of expression of plasmid genes. Quiescence would be a useful safeguard against loss of plasmids, which can cause the culture to be overgrown by plasmid-free bacteria. It could also help to reduce the frequency of undesirable mutations and provide a supplementary bio-safety mechanism, if escaping bacteria were unable to replicate.
Regulator of cell division RNA
Rcd is a functional RNA that is part of the native plasmid stability mechanisms. As Blaby and Summers 2009 describe, Rcd transcription is triggered by plasmid dimerisation and causes a delay in cell division. This allows the cell more time to resolve the dimer. This is important in passively distributed plasmids as it prevents dimer catastrophes. Sharpe et al. 1999 describes the structure of different forms of Rcd. The best one to use for our intent is probably Rcd70, which is the shortest isolated form.
Rcd does not have a recognizable terminator, but still seems to terminate properly. The RNA is unusually stable with a half-life of ~70 to 90 minutes. One problem encountered in culturing commercial Q-cells is that it is very difficult to fully stop the expression of Rcd. Small amounts of Rcd in the cell can slow the cell cycle down, creating an evolutionary incentive to become resistant to Rcd. Thus, cells that have been grown with a repressed Rcd gene, will lose the capability to become quiescent over time. Rowe and Summers 1999 achieved a functional quiescence system by putting the Rcd gene under a promoter repressed by a temperature sensitive form of Phage lambda cI repressor. This promoter system was the only one they could find that provided sufficient suppression.
h-ns mutants
This system of inducing quiescence only seems to work in specific h-ns mutant strains. H-NS is a nucleoid protein composed of a DNA binding and a regulatory domain, that usually acts as a transcriptional repressor. This book by NP Higgins has imformation on the properties of H-NS and its functions in the bacterial nucleoid. Mutants have a condensed chromosome, show higher expression of some genes, are non-motile and mucoidal, however the phenotypes are allele specific. We have been kindly provided with a selection of h-ns mutant strains by Dr Rowe.
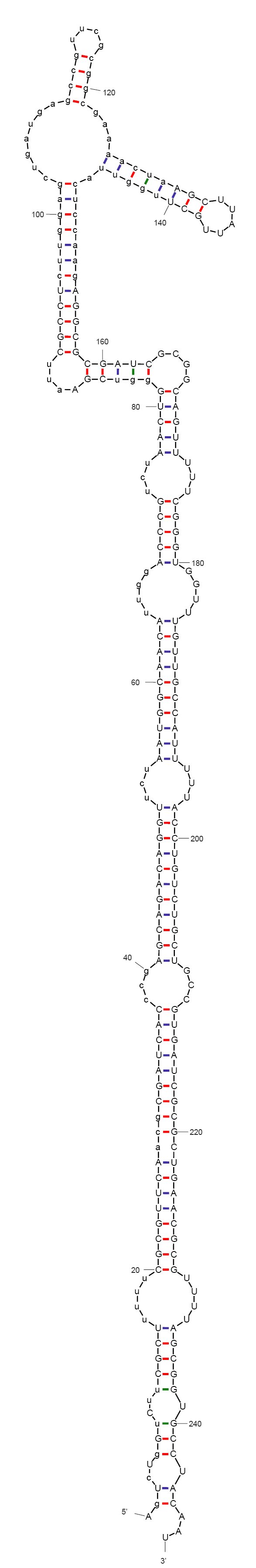
The aptamer-controlled-ribozyme approach
A new approach to making a quiescence switch would be to control the functionality of the folded RNA instead of its transcription, which proved to be problematic. Ideally the bacteria would grow in the lab or a vat in the presence of a ligand but would stop growing without dying, as soon as the ligand is removed. Thus if the engineered bacteria escaped into the wild they would stop growing very quickly as the ligand is diluted.
We hope to achieve this by rationally designing a gene for an RNA that folds into a non-functional shape. The structure will include a hammerhead ribozyme domain (see Martick and Scott 2006) that will hopefully cleave itself in vivo to release the functional Rcd RNA. In order to control this cleavage, the final construct would include an aptamer, which is an RNA structure able to bind a specific compound. This would create an allosteric ribozyme.
The project would involve synthesising different constructs (which are all mercifully short) and testing their functionality and sensitivity. The result would be a fairly short, but quite versatile BioBrick.
According to mFold and CLC workbench, the sequence we designed folds into a non-functional form, as expected. The predicted secondary structure is depicted on the left. This sequence does not include the aptamer yet. We are now investigating aptamers binding theophylline. This compound has been very described in the context of allosteric ribozymes, is easily available, non-toxic in low concentrations (naturally found in black tea), and non-native to bacteria.
Smolke et al. 2009 gives a nice review of aptamer activated hammerhead ribozymes and other functional RNA structures. More information on ribozymes (self- or trans-cleaving) can be found in Serganov and Patel 2007.
page summarising all RNA software
Intellectual Property
Using Rcd in the h-ns mutant to induce quiescence in E.coli is the Intellectual Property of Cambridge Microbial technologies. Dr Summers has been very supportive and we are going to continue the project. However, the legal issues have not been completely resolved yet. We still need to talk to Robert Hulme, the co-holder of the patent.
 "
"