Team:Calgary/Modelling/Maya
From 2010.igem.org
(Difference between revisions)
(New page: {{CalgaryMenu}} <html> <head> <style> div.container{ background-image:url("http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/PageContainer-1.png"); background-repeat: r...) |
|||
| (5 intermediate revisions not shown) | |||
| Line 44: | Line 44: | ||
#bodytitle h1{ | #bodytitle h1{ | ||
color:#54bcf8; | color:#54bcf8; | ||
| + | } | ||
| + | |||
| + | div.mainbody ol li{ | ||
| + | font-family:verdana; | ||
} | } | ||
| Line 61: | Line 65: | ||
<ul> | <ul> | ||
<li><a href="https://2010.igem.org/Team:Calgary/Modelling/MATLAB">MATLAB Models</a></li> | <li><a href="https://2010.igem.org/Team:Calgary/Modelling/MATLAB">MATLAB Models</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Calgary/Modelling/Maya">Maya | + | <li><a href="https://2010.igem.org/Team:Calgary/Modelling/Maya">Maya Animation</a></li> |
<ul> | <ul> | ||
| Line 70: | Line 74: | ||
<div class="mainbody"> | <div class="mainbody"> | ||
| - | <span id="bodytitle"><h1>Maya | + | <span id="bodytitle"><h1>Maya Animation</h1></span> |
| + | |||
| + | <p>As a side animation project, we are using <i>Maya</i>, a 3D graphics and animation program provided to us by Autodesk, who is a sponsor of iGEM this year. We intended to create an animation visualizing what the process of inclusion body formation might look like. The basis of this animation can be found in literature (Roberts, 2007).</p> | ||
| + | |||
| + | <img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/MayaFigure.png"></img> | ||
| + | |||
| + | <p>We believe this animation will help students in iGEM visualize and understand the process behind inclusion body formation. A dynamic visual catches people's attention and shows things as they happen, rather than showing static intermediate images on a diagram.</p> | ||
| + | |||
| + | <p>This is what we are showing: | ||
| + | <ol> | ||
| + | <li>A misfolded protein forms, exposing hydrophobic side chains. (0:00)</li> | ||
| + | <li>A nucleus of aggregation is formed by a collection of these misfolded proteins sticking together. (0:01)</li> | ||
| + | <li>More misfolded protein become attracted to the nucleus, attempting to "cover up" hydrophobic ends. (0:02 - 0:13)</li> | ||
| + | <li>Smaller clumps stick together into larger clumps. (0:14 - 0:23) | ||
| + | <li>This process continues until a large inclusion body forms in the cell, large enough to be visible. (0:23 - 0:38)</li> | ||
| + | </ol></p> | ||
| + | <br/> | ||
| - | + | <object width="480" height="385"><param name="movie" value="http://www.youtube.com/v/IouuHDjVpbo?fs=1&hl=en_US"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/IouuHDjVpbo?fs=1&hl=en_US" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="480" height="385"></embed></object> | |
| + | <br/> | ||
| + | <h2>References</h2> | ||
| + | <p>Roberts, C.J. (2007). Non-Native Protein Aggregation Kinetics. <i>Biotechnology and Bioengineering 98</i>(5), 927-938.</p> | ||
</div> | </div> | ||
Latest revision as of 01:07, 28 October 2010

Maya Animation
As a side animation project, we are using Maya, a 3D graphics and animation program provided to us by Autodesk, who is a sponsor of iGEM this year. We intended to create an animation visualizing what the process of inclusion body formation might look like. The basis of this animation can be found in literature (Roberts, 2007).
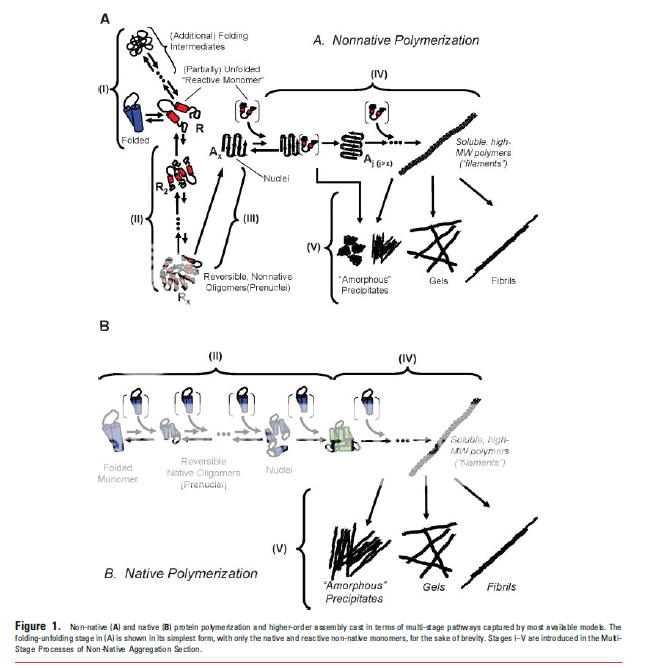
We believe this animation will help students in iGEM visualize and understand the process behind inclusion body formation. A dynamic visual catches people's attention and shows things as they happen, rather than showing static intermediate images on a diagram.
This is what we are showing:
- A misfolded protein forms, exposing hydrophobic side chains. (0:00)
- A nucleus of aggregation is formed by a collection of these misfolded proteins sticking together. (0:01)
- More misfolded protein become attracted to the nucleus, attempting to "cover up" hydrophobic ends. (0:02 - 0:13)
- Smaller clumps stick together into larger clumps. (0:14 - 0:23)
- This process continues until a large inclusion body forms in the cell, large enough to be visible. (0:23 - 0:38)
References
Roberts, C.J. (2007). Non-Native Protein Aggregation Kinetics. Biotechnology and Bioengineering 98(5), 927-938.
 "
"