Team:Calgary/30 July 2010
From 2010.igem.org
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| + | Friday July 30, 2010| | ||
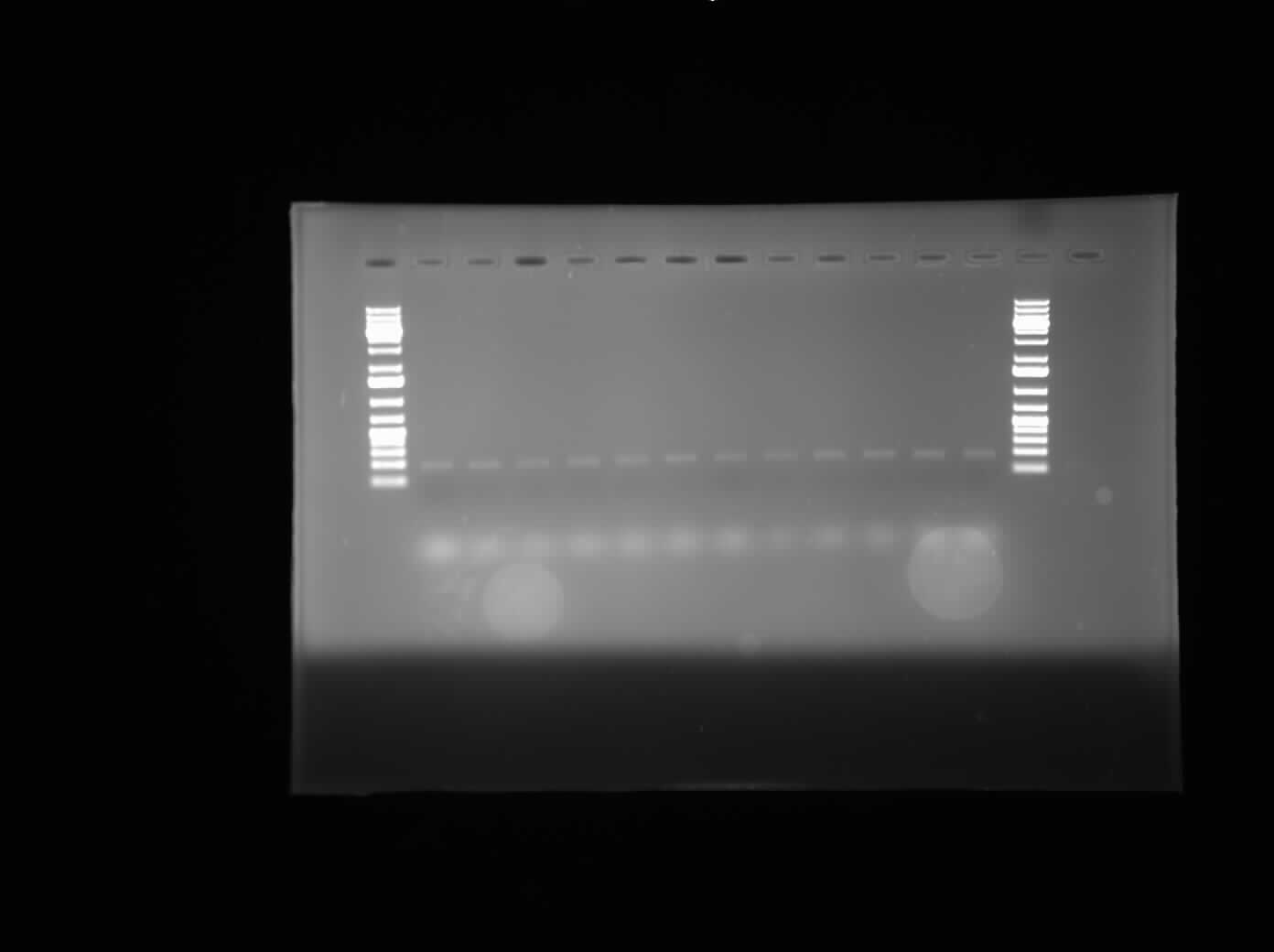
[[Image:07.30.2010-ChriscpxPgel.jpg|thumb|400px|Chris's first gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 1.5 µM.]] | [[Image:07.30.2010-ChriscpxPgel.jpg|thumb|400px|Chris's first gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 1.5 µM.]] | ||
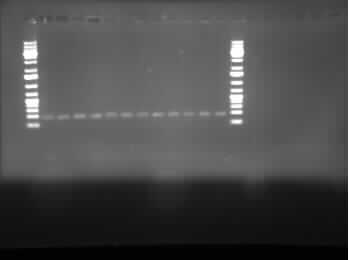
[[Image:07.30.2010-ChriscpxPgel2.jpg|thumb|400px|Chris's second gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 2.0 µM.]] | [[Image:07.30.2010-ChriscpxPgel2.jpg|thumb|400px|Chris's second gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 2.0 µM.]] | ||
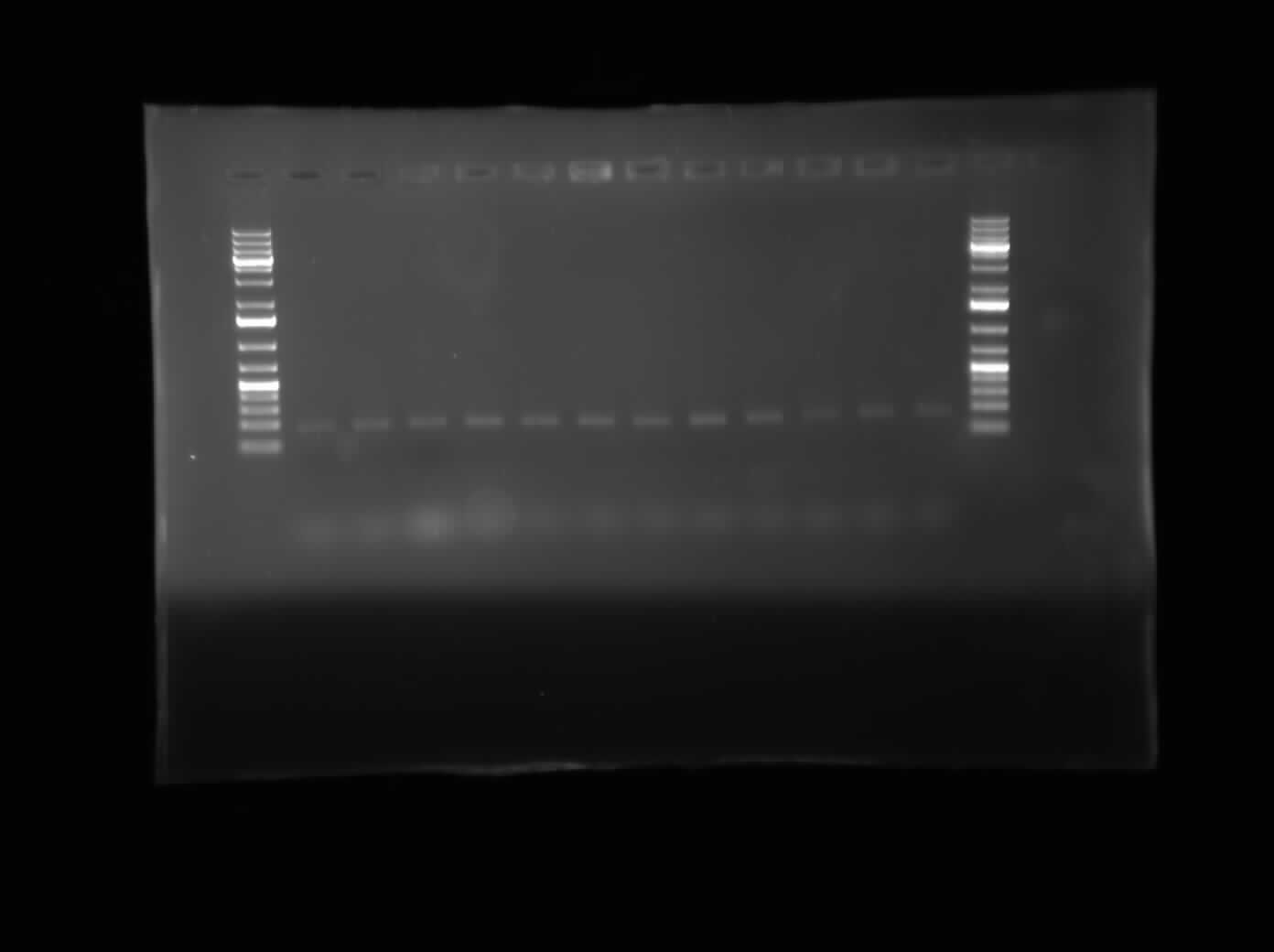
[[Image:07.30.2010-ChriscpxPgel3.jpg|thumb|400px|Chris's third gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 2.5 µM]] | [[Image:07.30.2010-ChriscpxPgel3.jpg|thumb|400px|Chris's third gel of the attempted gradient PCR to remove the CpxP promoter from the E. coli genome. This line of tubes ran from 50°C to 56°C and at a MgCl<sub>2</sub> concentration of 2.5 µM]] | ||
| - | |||
| - | |||
| - | |||
<u> Himika </u> | <u> Himika </u> | ||
| Line 27: | Line 25: | ||
[[Image:07.30.2010.JeremypRFPGradientSuccess copy.jpg|thumb|400px|Jeremy's Gradient PCR of biobricking pRFP. This used forward primer EcoRI, XbaI and gene specific region and reverse primer was gene specific and SpeI.]] | [[Image:07.30.2010.JeremypRFPGradientSuccess copy.jpg|thumb|400px|Jeremy's Gradient PCR of biobricking pRFP. This used forward primer EcoRI, XbaI and gene specific region and reverse primer was gene specific and SpeI.]] | ||
| + | |||
| + | |||
<u>Jeremy</u> | <u>Jeremy</u> | ||
Latest revision as of 04:53, 23 August 2010

Friday July 30, 2010
Himika
Today there was a meeting which updated updates all the team members about each other. I also kept looking into MATLAB tutorials and tried out some of the tutorials. I also read some of the papers about inclusion bodies. This will be used to model of the project. The sequence data of the I0500-B0034 came back today and it was successful it seems. So I have decided to go forth with the construct and add on more parts next week.
Chris
Today, I did the minipreps for the constructions of I0500-I13504 and I0500-I13507 using the Sigma Aldrich Plasmid Preparation kit. As well, I ran 3 gels of the gradient PCR which removed the CpxP promoter from the E. coli genome. The three different lines were mixed together so there are three tubes of PCR product instead of 36. Finally, Henry gave us instruments which would allow for the vacuum separation of PCR products from contaminents.
Emily
Today Raida and I ran a gel of our other two PCR's of malEdelSS and malE31delSS. They showed some amplification, however the bands were very faint. We set up two more and left them running overnight using different annealing temperatures. For one, we did 52 for 3 cycles, 53 for 3 cycles and then 54 for 3 cycles. For the other one, we put the annealing temperature much higher at 58. I will be coming in to run a gel of both of these PCR's tomorrow afternoon. Today we also had a meeting to clarify some of the criteris for iGEM and what needs to get donw in the next couple of days. Finally, I set up overnight cultures of malE and malE31 in TOPO in K broth. I will be coming in to miniprep these cultures tomorrow.
Patrick
Lots of Maya to still catch up on. Today I finished up the provided tutorial on polygon meshes. Next will be basic keyframe and motion path animation, as well as rendering. This will most likely be looked over during the weekend.
Jeremy
Today I did a gradient PCR of pRFP in the previous conditions that worked. And ran the gel at 0.8% at 90V. The conditions at 65.2 degrees celsius annealing temperature gave the best results. From this gel it seems that pRFP was successfully biobricked. I placed 5 PCR tubes in the column at 65.2 degrees so hopefully this will be enough to plasmid switch the product. I am also working various designs of buttons and tabs for the wiki. In the meeting with the supervisors, for the following week I am trying to plasmid switch pRFP, finish t-shirt designs as well as put up a template for the wiki.
 "
"