Team:Calgary/27 July 2010
From 2010.igem.org
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| - | + | Tuesday July 27, 2010| | |
| - | + | ||
| - | + | ||
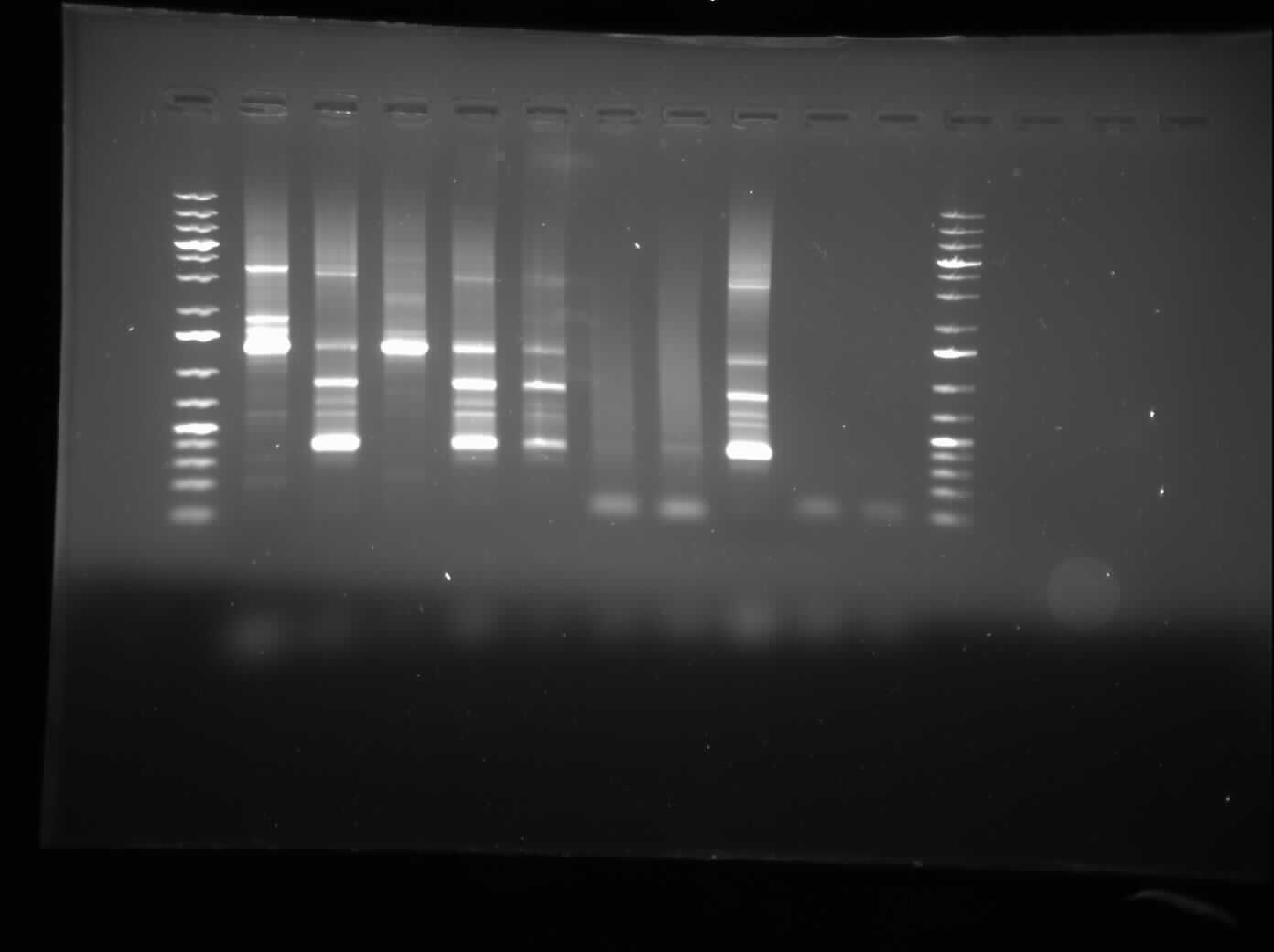
[[Image:07.27.2010-CpxPPCR.jpg|thumb|400px|Chris's gel electrophoresis of the plasmid PCR of the CpxP promoters.]] | [[Image:07.27.2010-CpxPPCR.jpg|thumb|400px|Chris's gel electrophoresis of the plasmid PCR of the CpxP promoters.]] | ||
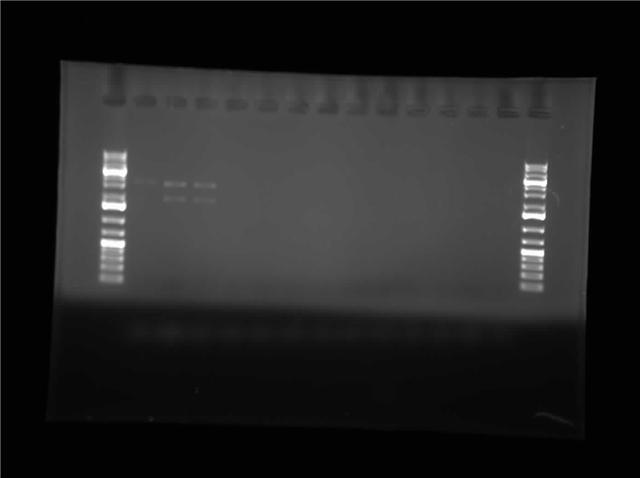
| + | [[Image:07.27.2010-Himika I0500-b0034 B1,C1 jpg.jpg|thumb|400px|Himika's gel of construction digest. I0500-B0034 and P1010-pSB1AK3]] | ||
| + | |||
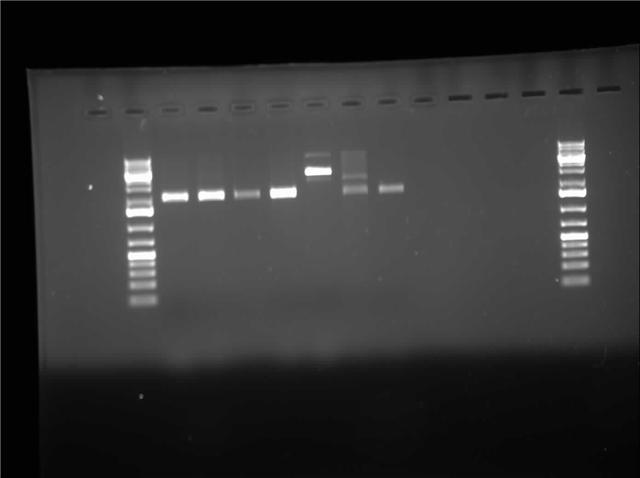
| + | [[Image:07.27.2010-Himika I0500-b0034-RD-pSB1A3.jpg|thumb|400px| Himika's gel of EcoRI and PstI digest from construction plasmid pSB1A3]] | ||
<u>Chris</u> | <u>Chris</u> | ||
| - | Today, I ran a 1.0% agarose gel electrophoresis of the CpxP promotor PCR that was left to be done overnight. | + | Today, I ran a 1.0% agarose gel electrophoresis of the CpxP promotor PCR that was left to be done overnight. The gel result showed a many bands all over the place, and will be redone to check. As well, we plan to re-PCR the CpxP promoter out of the genome. As well, overnight cultures were prepared and a construction from yesterday was finished. I talked to representatives from VWR and Corning Life Sciences about possible sponsorships and they agreed to provide our lab with materials that we need. |
| + | |||
<u>Jeremy</u> | <u>Jeremy</u> | ||
Today I ran a 0.8% gel. Prepared overnights for the I0500+B0034 in AK ccdB. | Today I ran a 0.8% gel. Prepared overnights for the I0500+B0034 in AK ccdB. | ||
| + | |||
| + | |||
| + | <u>Patrick</u> | ||
| + | |||
| + | We have decided to start using Autodesk Maya to create an animation of inclusion body formation. This means I need to ''learn'' Autodesk Maya. Thus, I spent much of the day doing so. Jeremy and I also discussed the things we need the rest of the team to help contribute to the wiki. | ||
| + | |||
| + | |||
| + | <u>Emily</u> | ||
| + | |||
| + | Unfortunately the gel of my PCR to Biobrick malE and malE31 showed no amplification whatsoever so I have to start this again. Today I did a gradient PCR to try to biobrick malEdelSS and malE31delSS. They are currently in the plasmids supplied to us by the Betton Labs. I also set up two more gradient PCRs to try to biobrick malE and malE31. I also set up a restriction digest with XbaI and SpeI of the PCR product from the attempt at biobricking malE and malE31 from last week. I left these to digest overnight along with psb1AC3 and psb1AK3 vectors in order to attempt to get biobricked malE and malE31 into the BBK vectors. I will transform these tomorrow and then plate them for overnight growth. I also looked into a paper on malE and the cpxP, cpxR and degP pathways. | ||
| + | |||
| + | |||
| + | <u>Himika</u> | ||
| + | |||
| + | Today I retried plasmid switch of I0500-B0034. The difference this time is that I used P1010 in pSB1AK3. So I am expecting cells to grow tomorrow which consist of the insert. I also ran a restriction digest of I0500-B0034 in pSB1A3. I digested with EcoRI and PstI. This digest gave me gel with 1 band per lane. I will wait and see if the plates grow. If the cells fail to grow I will try retransforming again. I also did research on factors that might induce inclusion body formation. This information will be used to create a mathematical model in matlab. | ||
| + | |||
}} | }} | ||
Latest revision as of 04:51, 23 August 2010

Tuesday July 27, 2010
Chris
Today, I ran a 1.0% agarose gel electrophoresis of the CpxP promotor PCR that was left to be done overnight. The gel result showed a many bands all over the place, and will be redone to check. As well, we plan to re-PCR the CpxP promoter out of the genome. As well, overnight cultures were prepared and a construction from yesterday was finished. I talked to representatives from VWR and Corning Life Sciences about possible sponsorships and they agreed to provide our lab with materials that we need.
Jeremy
Today I ran a 0.8% gel. Prepared overnights for the I0500+B0034 in AK ccdB.
Patrick
We have decided to start using Autodesk Maya to create an animation of inclusion body formation. This means I need to learn Autodesk Maya. Thus, I spent much of the day doing so. Jeremy and I also discussed the things we need the rest of the team to help contribute to the wiki.
Emily
Unfortunately the gel of my PCR to Biobrick malE and malE31 showed no amplification whatsoever so I have to start this again. Today I did a gradient PCR to try to biobrick malEdelSS and malE31delSS. They are currently in the plasmids supplied to us by the Betton Labs. I also set up two more gradient PCRs to try to biobrick malE and malE31. I also set up a restriction digest with XbaI and SpeI of the PCR product from the attempt at biobricking malE and malE31 from last week. I left these to digest overnight along with psb1AC3 and psb1AK3 vectors in order to attempt to get biobricked malE and malE31 into the BBK vectors. I will transform these tomorrow and then plate them for overnight growth. I also looked into a paper on malE and the cpxP, cpxR and degP pathways.
Himika
Today I retried plasmid switch of I0500-B0034. The difference this time is that I used P1010 in pSB1AK3. So I am expecting cells to grow tomorrow which consist of the insert. I also ran a restriction digest of I0500-B0034 in pSB1A3. I digested with EcoRI and PstI. This digest gave me gel with 1 band per lane. I will wait and see if the plates grow. If the cells fail to grow I will try retransforming again. I also did research on factors that might induce inclusion body formation. This information will be used to create a mathematical model in matlab.
 "
"