Team:Calgary/14 June 2010
From 2010.igem.org

Monday June 14, 2010
Dev, Chris, and Jeremy
Today, we did the constructions of J13002 (a combination of a constitutive promoter with a RBS) and E0040 (GFP) and E0032 (YFP) to determine if the two fluorescent proteins were functional. As well, a plasmid switch was begun with the part J23032 into a Kan plasmid backbone so a selection pressure would be present when it was constructed with R0040. This construction was done using standard Biobrick cloning and then transformation into Top10 Competent cells. We received the parts from the registry in the form of agar stabs on friday June 11, 2010 but they were received too late to work with them. We also did a construction of B0015 to R0040 to begin the RNA detection circuit. The agar stabs were streaked onto plates and left to grow overnight of the parts I0500, K239000, K135000, and K274210.
Alex, Patrick, and Raida
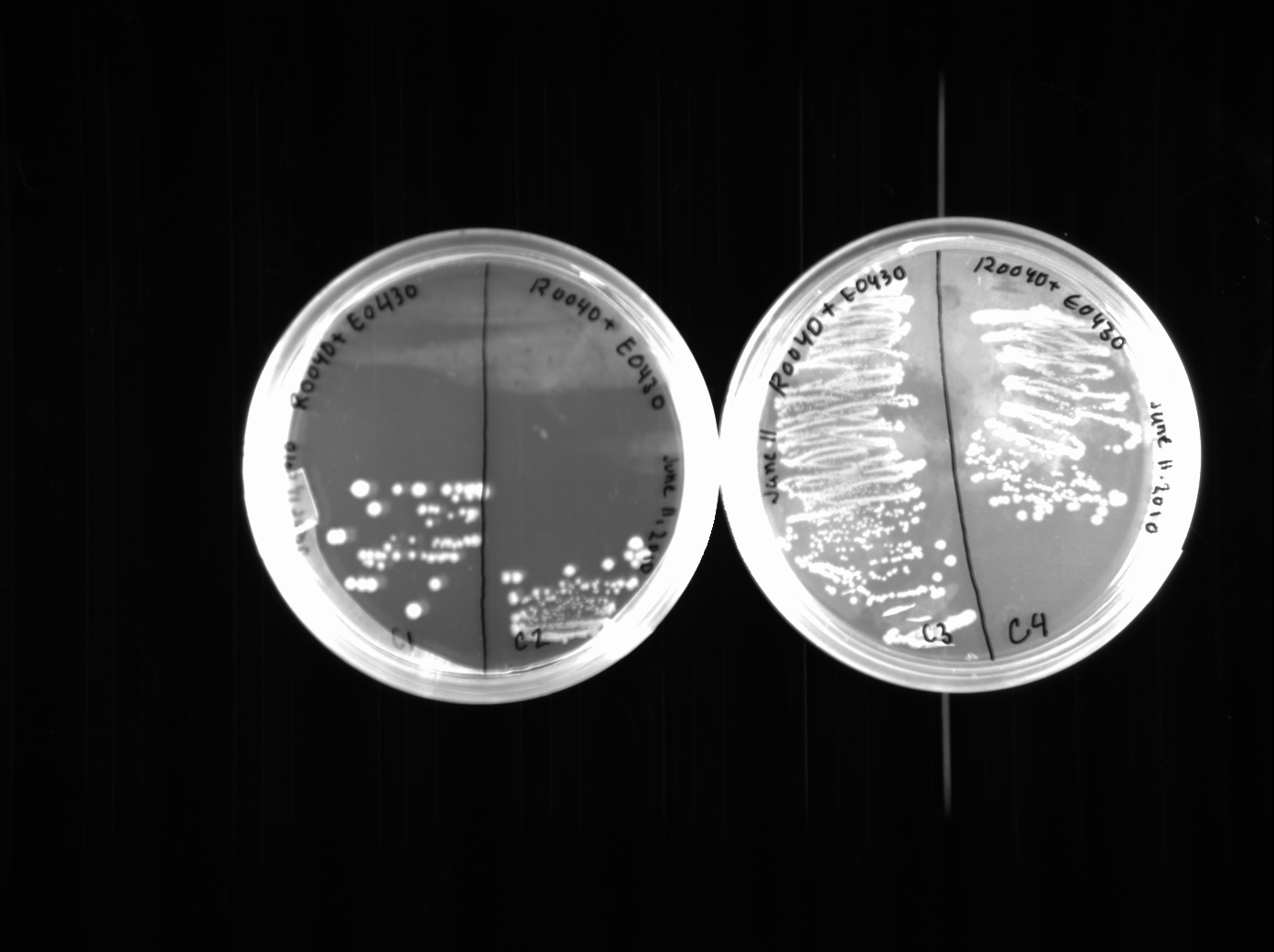
Surprisingly, we seemed to have picked good colonies because three out of four restreaks were distinctively yellow. The colonies are quite large, though, since we left them growing over the weekend. We did a colony PCR of these three restreaks, though the results were not very clear, nor very impressive. We will try and do another colony PCR when we finish switching and transforming the E0430 plasmid, which we also did today. We switched the E0430 (from colonies 3 and 4) to a pSB1K2 plasmid. Each was plated onto two kanamycin plates.
As well, we also did an agar stab of the K239000 (DegP promoter) that arrived last Friday. This was streaked onto two ampicillin plates. It was discovered that the promoter was not in a normal BioBrick plasmid, but it is still likely that the BioBrick prefix and suffix are still in place, meaning we likely still able to digest the plasmid with EcoRI and PstI to switch the part into some other plasmid. Finally, we also transformed the E0420 (ECFP generator with no LVA tag) into cells.
Emily
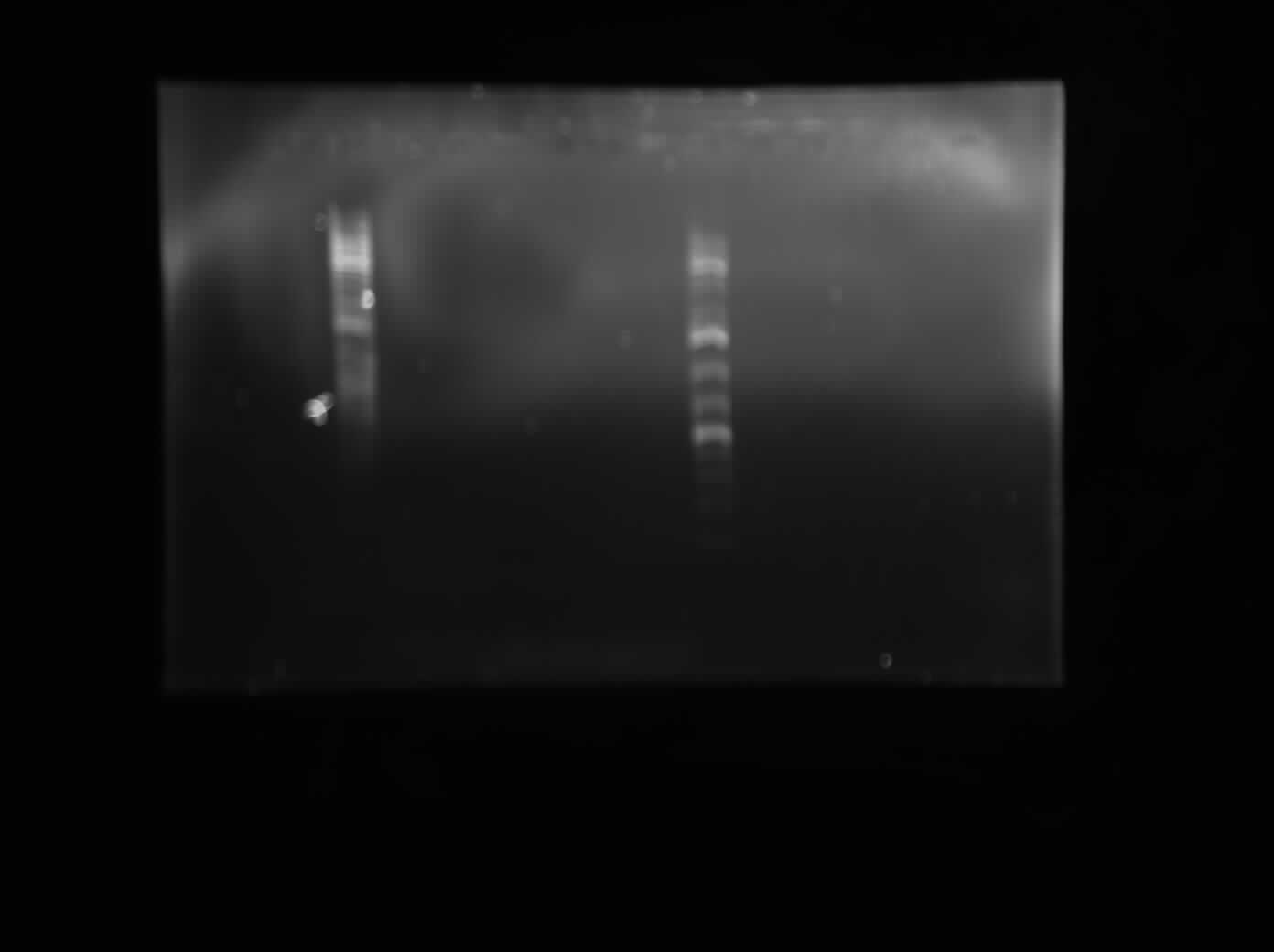
Today I did a restriction digest of all of the Biobrick vectors that Chris transformed form the registry, just to make sure that they contain all of the Biobrick cut sites. I digested each vector (psB1K2, psB1A2, psB1AC3 and psB1AK3) with each combination of construction restriction enzymes (EcoRI+SpeI, EcoRI+PstI, XbaI+PstI and XbaI+SpeI). I also digested my two colonies of my 10500-B0034 construct (colonies 6+7) with EcoRI and SpeI. I left these digest for two hours and then I ran all of them on a 1% agarsoe gel with 10x Orange Loading dye and ddH20. The gel is pictured below. From this, we concluded that all of the vecotrs have the expected Biobrick cut sites. Also, we saw bands of expected sizes in the I0500-B0034 digest, although the presence of a third, much larger band indicates that the digestion was perhaps not a cmplete digest but rather a partial digest and that some of the undigested product is still present. More verification will be done on these colonies before they are sent down for sequencing. Also, a positive control was not run for this experiment, which should be done next time.
Himika
Today, I did a restriction digest for the parts E1010, B0034 and the B0034-E1010 construct with Biobrick restriction enzymes to verify the size of the parts and to see if the construct worked. Once again the gel gave no bands but this time it was determined the TAE buffer was not working well and the DNA was probably floating out of the wells. As a result of this we made another 1xTAE buffer and the experiment will be repeated later. As well, I did a construction of the second half of the circuit of the ibpAB-fsx fusion promoter which consists of the E1010-B0015 reporter terminator circuit.
 "
"