Team:Alberta/Building Parts
From 2010.igem.org
| (54 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | |||
| + | <head> | ||
| + | |||
| + | <style type="text/css"> | ||
| + | |||
| + | /* GETS RID OF DEFAULT TITLE */ | ||
| + | .firstHeading {display:none;} | ||
| + | |||
| + | |||
| + | /* testing different titles css for bios */ | ||
| + | |||
| + | img.float-left | ||
| + | { | ||
| + | float:left; display:inline; margin-right:10px; | ||
| + | } | ||
| + | |||
| + | /* SETS OUTSIDE BGCOLOUR AND INSIDE BGCOLOUR */ | ||
| + | |||
| + | body | ||
| + | { | ||
| + | background-color: #252525; | ||
| + | margin: 0px; | ||
| + | padding: 0px; | ||
| + | border: 0px; | ||
| + | font-family:"Trebuchet MS", arial; | ||
| + | } | ||
| + | |||
| + | #title | ||
| + | { | ||
| + | background: #ffffff; | ||
| + | width:28.5%; | ||
| + | float: left; | ||
| + | padding: 0em; | ||
| + | font-size: 3.4em; | ||
| + | color: #757575; | ||
| + | height: 2em; | ||
| + | margin-left:1.5%; | ||
| + | vertical-align: middle; | ||
| + | } | ||
| + | |||
| + | #navbar | ||
| + | { | ||
| + | background: #ffffff; | ||
| + | width:65.5%; | ||
| + | float: right; | ||
| + | padding: 0em; | ||
| + | text-align:right; | ||
| + | font-size: 1em; | ||
| + | color: #a5a5a5; | ||
| + | margin-right:2.5%; | ||
| + | vertical-align: middle; | ||
| + | height: 2.5em; | ||
| + | display: inline; | ||
| + | } | ||
| + | |||
| + | #highlight | ||
| + | { | ||
| + | background: #fff200; | ||
| + | width:100%; | ||
| + | clear: both; | ||
| + | padding: 0em; | ||
| + | height: 3px; | ||
| + | } | ||
| + | |||
| + | #photo | ||
| + | { | ||
| + | width: 965px; | ||
| + | float: left; | ||
| + | padding: 0em; | ||
| + | height: 200px; | ||
| + | /* height: 200px; */ | ||
| + | background: url(https://static.igem.org/mediawiki/2010/b/b6/Alberta_GroupPic.png); | ||
| + | background-position: 0% 0%; | ||
| + | border: 0em; | ||
| + | } | ||
| + | |||
| + | p.logo | ||
| + | { | ||
| + | vertical-align: top; | ||
| + | } | ||
| + | |||
| + | .llink2 A:link {text-decoration: underline; color: #757575; font-size: 1em;} | ||
| + | .llink2 A:visited {text-decoration: none; color: #656565; font-size: 1em;} | ||
| + | .llink2 A:hover {text-decoration: none; font-size: 1em;} | ||
| + | .llink2 A:active {text-decoration: none; font-size: 1em;} | ||
| + | |||
| + | #content | ||
| + | { | ||
| + | /*width:69.5%;*/ | ||
| + | height:50%; | ||
| + | border:1px solid #000000; | ||
| + | } | ||
| + | |||
| + | # content p | ||
| + | { | ||
| + | font-family:"Trebuchet MS", arial; | ||
| + | font-size: 1em; | ||
| + | color: #000000; | ||
| + | } | ||
| + | #highlightb | ||
| + | { | ||
| + | background: #ffffff; | ||
| + | width:100%; | ||
| + | clear: both; | ||
| + | padding: 0em; | ||
| + | height: 3px; | ||
| + | } | ||
| + | |||
| + | #blacktop | ||
| + | { | ||
| + | background: #3b3b3b; | ||
| + | width: 100%; | ||
| + | clear: both; | ||
| + | padding: 0em; | ||
| + | height: 1.3em; | ||
| + | text-align: center; | ||
| + | font-family: "Trebuchet MS", Times; | ||
| + | color: #fff200; | ||
| + | font-size: 2.5em; | ||
| + | line-height: 120%; | ||
| + | border-top: 0.1em solid #ffffff; | ||
| + | } | ||
| + | |||
| + | #footer | ||
| + | { | ||
| + | clear: both; | ||
| + | margin: 0; | ||
| + | padding: .5em; | ||
| + | color: #333333; | ||
| + | background-color: #dddddd; | ||
| + | border-top: 3px solid #333333; | ||
| + | } | ||
| + | |||
| + | .llink3 A:link {text-decoration: underline; color: #757575; font-size: 1em;} | ||
| + | .llink3 A:visited {text-decoration: none; color: #656565; font-size: 1em;} | ||
| + | .llink3 A:hover {text-decoration: none; font-size: 1em;} | ||
| + | .llink3 A:active {text-decoration: none; font-size: 1em;} | ||
| + | |||
| + | .h1 { text-decoration: underline; color: #6a7a69; font-size: 2.5em;} | ||
| + | .h2 { margin-top: 8em; margin-bottom: 3em; color: #fff200; font-size: 2em;} | ||
| + | .h3 { margin-top: 8em; margin-bottom: 3em; color: #fff200; font-size: 2em;} | ||
| + | .h4 { margin-top: 8em; margin-bottom: 3em; color: #fff200; font-size: 2em;} | ||
| + | .h5 { margin-top: 8em; margin-bottom: 3em; color: #fff200; font-size: 2em;} | ||
| + | |||
| + | #vtab0 | ||
| + | { | ||
| + | width:20%; | ||
| + | float:left; | ||
| + | } | ||
| + | |||
| + | #vtab0 li | ||
| + | { | ||
| + | list-style:none; | ||
| + | background-color: #a5a5a5; | ||
| + | border:1px dotted #3b3b3b; | ||
| + | font-family: "Trebuchet MS", Times; | ||
| + | font-weight:bold; | ||
| + | font-size: 1em; | ||
| + | color: #ffffff; | ||
| + | } | ||
| + | |||
| + | #vtab0 li:hover | ||
| + | { | ||
| + | background-color: #3b3b3b; | ||
| + | } | ||
| + | |||
| + | #tabContent | ||
| + | { | ||
| + | float:left; | ||
| + | width:78%; | ||
| + | margin: 1% 1%; | ||
| + | font-family: "Trebuchet MS", Times; | ||
| + | font-style: bold; | ||
| + | font-size: 1em; | ||
| + | color: #000000; | ||
| + | } | ||
| + | |||
| + | .vtInfo | ||
| + | { | ||
| + | display:none; | ||
| + | height:98%; | ||
| + | overflow: auto; | ||
| + | } | ||
| + | |||
| + | .llink1 A:link {text-decoration: none; color: #bfbfbf; font-size: 1em;} | ||
| + | .llink1 A:visited {text-decoration: none; color: #bfbfbf; font-size: 1em;} | ||
| + | .llink1 A:hover {text-decoration: none; color: #fff200; font-size: 1em;} | ||
| + | .llink1 A:active {text-decoration: none; font-size: 1em;} | ||
| + | |||
| + | |||
| + | .llink3 A:link {text-decoration: underline; color: #757575; font-size: 1em;} | ||
| + | .llink3 A:visited {text-decoration: none; color: #656565; font-size: 1em;} | ||
| + | .llink3 A:hover {text-decoration: none; font-size: 1em;} | ||
| + | .llink3 A:active {text-decoration: none; font-size: 1em;} | ||
| + | |||
| + | .h2 {text-decoration: none; color: #757575; font-size: 2.5em; font-family: "Trebuchet MS", Times; line-height: 120%;} | ||
| + | .h3 {text-decoration: none; color: #757575; font-size: 1em; font-family: "Trebuchet MS", Times;} | ||
| + | |||
| + | </style> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | |||
| + | $(document).ready( function(){ | ||
| + | // load group photo on page load | ||
| + | $('#vtab1').show(); | ||
| + | |||
| + | |||
| + | }); | ||
| + | |||
| + | function showTab(IDS) | ||
| + | { | ||
| + | var tabs = document.getElementById('tabContent').getElementsByTagName('div'); | ||
| + | var tabs = getElementsByClass('vtInfo',document.getElementById('tabContent'),'div'); | ||
| + | |||
| + | for (var i=0; i<tabs.length; i++) { tabs[i].style.display = 'none'; } | ||
| + | document.getElementById(IDS).style.display = 'block'; | ||
| + | } | ||
| + | |||
| + | function getElementsByClass(searchClass,node,tag) | ||
| + | { | ||
| + | var classElements = new Array(); | ||
| + | if ( node == null ) | ||
| + | node = document; | ||
| + | if ( tag == null ) | ||
| + | tag = '*'; | ||
| + | var els = node.getElementsByTagName(tag); | ||
| + | var elsLen = els.length; | ||
| + | var pattern = new RegExp("(^|\\s)"+searchClass+"(\\s|$)"); | ||
| + | for (i = 0, j = 0; i < elsLen; i++) { | ||
| + | if ( pattern.test(els[i].className) ) { | ||
| + | classElements[j] = els[i]; | ||
| + | j++; | ||
| + | } | ||
| + | } | ||
| + | return classElements; | ||
| + | } | ||
| + | |||
| + | </script> | ||
| + | |||
| + | |||
| + | </head> | ||
| + | |||
| + | |||
| + | <!-- ACTUAL BODY STARTS HERE ********************************** --> | ||
| + | <body> | ||
| + | |||
| + | <!-- PROJECT TITLE --> | ||
| + | |||
| + | <div id="title"> | ||
| + | |||
| + | <p class="logo"><img src="https://static.igem.org/mediawiki/2010/c/cc/Alberta_Logo.png" width="50px" height="50px" padding="5px" align="left"></img></p> | ||
| + | |||
| + | genomikon</div> | ||
| + | |||
| + | <!-- NAVBAR --> | ||
| + | <div id="navbar"><span class="llink2"><a href="https://2010.igem.org/Team:Alberta/JTest"> home</a> | <a href="project"> project</a> | <a href="ethics"> ethics</a> | <a href="parts"> parts</a> | <a href="software"> software</a> | <a href="https://2010.igem.org/Team:Alberta/Notebook"> notebook</a> | <a href="team"> team</a></span> | ||
| + | </div> | ||
| + | |||
| + | <!-- HIGHLIGHT --> | ||
| + | <div id="highlight"></div> | ||
| + | |||
| + | <div id="photo"></div> | ||
| + | |||
| + | <div id="highlightb"></div> | ||
| + | |||
| + | <br style="clear:both"> | ||
| + | </div> | ||
| + | |||
| + | <div id="Content"> | ||
| + | |||
| + | <div id="vtab0"> | ||
| + | <li onclick="showTab('vtab2')">Base Plasmids v.1</li> | ||
| + | <li onclick="showTab('vtab3')">Base Plasmids v.2</li> | ||
| + | <li onclick="showTab('vtab4')">Antibiotic Markers</li> | ||
| + | <li onclick="showTab('vtab5')">Abandoned Projects</li> | ||
| + | </div> | ||
| + | |||
| + | <div id="tabContent"> | ||
| + | |||
| + | <div id="blacktop">Building Parts</div> | ||
| + | |||
| + | <div id="vtab2" class="vtInfo"><span class="h2">Base Plasmids v.1</span> | ||
| + | |||
| + | <p>Our original base plasmids contain a Kanamycin resistance cassette (p1003) bracketed by BsaI or BfuAI or BbsI cut sites. If cut with either BsaI or BfuAI or BbsI, the Kanamycin cassette is release with sticky ends characteristic of an A or a B BioByte. </p> | ||
| + | |||
| + | <p><b>May 10, 2010</b> PCRed p1003 (Kanamycin cassette)with primers PrA_p1003+ and PrB'_p1003-</p> | ||
| + | <p><b>May 10, 2010</b> PCRed p1003 (Kanamycin cassette)with primers PrB_p1003+ and PrA'_p1003-</p> | ||
| + | <p><b>May 19, 2010</b> Digested pSB1C3 with Not1 </p> | ||
| + | <p><b>May 19, 2010</b> Digested PCR products of p1003 (Kanamycin cassette) Not1 </p> | ||
| + | <p><b>May 25, 2010</b> Ligated PCR products of p1003 (Kanamycin cassette) and pSB1C3 </p> | ||
| + | </div> | ||
| + | |||
| + | <div id="vtab3" class="vtInfo"><span class="h2">Testing Parts</span> | ||
| + | <p>Before we were able to test parts we created 2 base testing plasmids (vector 01 and vector 02). Vector 01 is designed to test Open Reading Frame parts, or parts that code for proteins. The part is flanked by a promoter and the start codon on one side and a stop codon and terminator on the other. Vector 02 is designed to test linker parts, or parts that control the expression of the Open Reading Frame parts they are next two. In Vector 02 the part is flanked by two distinct reporter genes, that by comparing the relative expression of the 2 reporter genes we can determine the behavior of the linking part.</p> | ||
| + | </div> | ||
| + | |||
| + | <div id="vtab4" class="vtInfo"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/b/b1/Alberta_Oscar.png" width="400px" height="533px" padding="0px" align="left"></img> | ||
| + | <span class="h2">Oscar Cortes</span><br> | ||
| + | <span class="h3">Specialized in Molecular Genetics (Graduate)</span> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div id="highlightb"></div> | ||
| + | |||
| + | <div id="footer"><span class="llink3"> | ||
| + | <a href="Site_Map">site map</a>|<a href="Sponsors">sponsors</a>|<a href="Contact">contact us</a> | ||
| + | </span></div> | ||
| + | |||
| + | </body> | ||
| + | |||
| + | </html> | ||
| + | |||
== Building Parts == | == Building Parts == | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | PCR | + | __notoc__ |
| - | Image of Gel | + | |
| + | ==10-05-2010== | ||
| + | |||
| + | <p>PCR a Kanamycin resistance cassette fragment from p1003 with primers containing either the A/B' ends or the B/A' ends. (Fragments formed called KanR A/B'-Bsa and KanR B/A'-Bsa respectively)</p> | ||
| + | <p>Recipe: | ||
| + | :1μL p1003 (approx. 1ng) | ||
| + | :2.5μL prA_p1003+ | ||
| + | :2.5μL prB'_p1003- | ||
| + | :5μL 10X PCR buffer | ||
| + | :1μL 10uM dNTPs | ||
| + | :2μL 50uM MgCl<sub>2</sub> | ||
| + | :0.5μL Taq polymerase | ||
| + | :35.5μL MilliQ H<sub>2</sub>O</p> | ||
| + | </br> | ||
| + | <p>Same recipe for KanR B/A'-Bsa except primers are prB_p1003+ and prA'_p1003-. </p> | ||
| + | <p>Program: | ||
| + | # 5 min-94<sup>o</sup>C | ||
| + | # 45 sec-94<sup>o</sup>C | ||
| + | # 1 min-60<sup>o</sup>C | ||
| + | # 1 min-72<sup>o</sup>C | ||
| + | # Repeat 2 through 4 35 times | ||
| + | # 5 min-72<sup>o</sup>C</p> | ||
| + | <!--Image of gel performed that day. 5μL of each PCR reaction, 1μL of 10X loading dye and 4μL MilliQ water in a 1% agarose gel --> | ||
| + | |||
| + | ==11-05-2010== | ||
| + | <p>PCR purification of KanR A/B'-Bsa and KanR B/A'-Bsa created [[#10-05-2010|10-05-2010]] with Qiagen PCR cleanup kit.</p> | ||
| + | <p>Determined concentrations by nanodrop. KanA/B'-Bsa: 101.1ng/μL KanB/A'-Bsa:89.6ng/μL</p> | ||
| + | |||
| + | ==17-05-2010== | ||
| + | <p>Innoculated 250mL overnight cultures with 10mL,4mL and 2mL of a starter culture of DH5α. Left shaking at 18<sup>o</sup>C overnight. </p> | ||
| + | |||
| + | ==18-05-2010== | ||
| + | <p>Prepared DH5α E.Coli competent cells using the Inoue Method. </p> | ||
| + | <p>Transformed DH5α cells with pSB1C3-J04450 and grew overnight at 37<sup>o</sup>C on Chloramphenicol plates</p> | ||
| + | |||
| + | ==19-05-2010== | ||
| + | <p>From the transformation of DH5α cells with pSB1C3-J04450 performed on [[#18-05-2010|18-05-2010]], we took 4 distinct colonies, streaked them on a new chloramphenicol plate and inoculated 5ml liquid cultures.</p> | ||
| + | |||
| + | ==20-05-2010== | ||
| + | <p>Performed a Miniprep of 3 of the 4 5ml liquid cultures of DH5α cells with pSB1C3-J04450 from [[#19-05-2010|19-05-2010]]. Took a 1μL sample of the Miniprep solutions and digested with NotI at 37<sup>o</sup>C for 1 hour. </p> | ||
| + | <p>Digestion Recipe: | ||
| + | :1μL Miniprep (between 153.2 ng/μl and 302.7ng/μl determined by nanodrop) | ||
| + | :1μL NotI | ||
| + | :1μL 10X ReACT 3 | ||
| + | :7μL MilliQ</p> | ||
| + | <p>Ran Digestion on a 1% agarose gel to check that the plasmid obtained with what we expected. </p> | ||
| + | <!-- Image of Gel--> | ||
| + | |||
| + | ==25-05-2010== | ||
| + | <p>Made 1.5mL LB liquid cultures of pSB1C3 from the plate streaked on [[#19-05-2010|19-05-2010]] and added chloramphenicol.</p> | ||
| + | |||
| + | ==26-05-2010== | ||
| + | <p>Made 3 glycerol stocks of pSB1C3 from overnight made [[#25-05-2010|25-05-2010]]</p> | ||
| + | |||
| + | ==27-05-2010== | ||
| + | </p>Digested both A/B' and B/A' Kanamycin Resistance cassettes fragments from [[#11-05-2010|11-05-2010]] and pSB1C3 from [[#20-05-2010|20-05-2010]] with NotI at 37<sup>o</sup>C for 1 hour. Heat inactivated the NotI for 10 minutes at 65<sup>o</sup>C. Ligated the Kanamycin Resistance cassettes into pSB1C3 at 16<sup>o</sup>C for 1 hour then took 15μL to room temperature for 2 hours. Transformed 100μL of DH5α cells with 5μL of RT ligation reaction. Plated transformation on plates with both Chloramphenicol and Kanamycin.</p> | ||
| + | <p>Digestion Recipe: | ||
| + | :1μL Miniprep (302.7ng/μl determined by nanodrop) | ||
| + | :2μL either A/B' or B/A' Kanamycin resistance cassette (approx. 100ng/μL) | ||
| + | :1μL NotI | ||
| + | :1μL 10X ReACT 3 | ||
| + | :5μL MilliQ</p> | ||
| + | <p>Ligation Recipe: | ||
| + | :10μL of Digest solution | ||
| + | :1μL T4 DNA ligase | ||
| + | :6μL 5X Buffer | ||
| + | :13μL MilliQ H<sub>2</sub>O</p> | ||
| + | <p>Also transformed pSB4A5-J04450, pSB4C5-J04450 and pSB3T5-J04450 from the 2010 biobrick parts into DH5α cells.</p> | ||
| + | <p>Performed PCR reactions to create parts with antibiotic resistance with negative controls.</p> | ||
| + | <p>PCR Recipe: | ||
| + | :3μL 10X PCR Buffer | ||
| + | :1μL 10 uM dNTPs | ||
| + | :2μL 50 uM MgCl<sub>2</sub> | ||
| + | :17.5μL MilliQ H<sub>2</sub>O | ||
| + | :0.5μL Taq Polymerase | ||
| + | :1μL Template (psB4A5-J04450, psB4C5-J04450 or psB3T5-J04450) | ||
| + | :2.5μL Primer + (PrA psB4A5 ApR+, PrA psB4C5 ChR+ or PrA psB3T5 TR+) | ||
| + | :2.5μL Primer - (PrB psB4A5 ApR-, PrB psB4C5 ChR- or PrB psB3T5 TR-)</p> | ||
| + | <p>PCR Program: | ||
| + | # 5 min-94<sup>o</sup>C | ||
| + | # 45 sec-94<sup>o</sup>C | ||
| + | # 1 min-60<sup>o</sup>C | ||
| + | # 1 min-72<sup>o</sup>C | ||
| + | # Repeat 2 through 4 35 times | ||
| + | # 5 min-72<sup>o</sup>C</p> | ||
| + | <!--gel images of PCR Products (Alina's) and ligated and pre-digested samples (Jeremy's)--> | ||
| + | |||
| + | ==28-05-2010== | ||
| + | <p>We got colonies!! (it's a fantastic feeling) We then lovingly put them in the cold room to await our return from Calgary</p> | ||
| + | |||
| + | ==30-05-2010== | ||
| + | <p>From the transformation of DH5α cells with pSB1C3-KanR performed on [[#28-05-2010|28-05-2010]], we took 12 distinct colonies of each KanA/B' and KanB/A', streaked them on a new chloramphenicol plate and inoculated 5ml liquid cultures with the appropriate antibiotics overnight at 37<sup>o</sup>C. We also picked colonies of pSB4A5-J04450, pSB4C5-J04450 and pSB3T5-J04450, streaked and made 5mL liquid cultures of them too.</p> | ||
| + | |||
| + | ==31-05-2010== | ||
| + | <p>9/12 of the pSB1C3-KanA/B' Liquid cultures [[#30-05-2010|30-05-2010]] were successful and only 1/12 of the pSB1C3-KanB/A' liquid cultures were successful. The pSB4A5, pSB3T5 and pSB4C5 liquid cultures worked. Miniprepped all the liquid cultures that worked. However, the streaks on the plates worked.</p> | ||
| + | <p>Performed a restriction digest of an aliquot of the pSB1C3-KanA/B' and pSB1C3-KanB/A' minipreps. Digested with XbaI at 37<sup>o</sup>C for one hour and then with EcoRI at 37<sup>o</sup>C for one hour. Ran a 1% agarose gel of the digests to determine the orientation of the KanR fragments. </p> | ||
| + | <!--who did this digest!! we need an image--> | ||
| + | <p>KanA/B' and KanB/A' fragements PCRed on [[#11-05-2010|11-05-2010]], digested with BsaI at 37<sup>o</sup>C for 1.5hours, heat inactivated at 65<sup>o</sup>C for 30 minutes. Tried to ligate KanA/B' fragments to each other and tried to ligate KanB/A' fragments to each other. Also tried to ligate KanA/B' fragments with KanB/A'. Ligated with T4 DNA ligase overnight at 16<sup>o</sup>C.</p> | ||
| + | |||
| + | ==01-06-2010== | ||
| + | |||
| + | KanA/B' and KanB/A' fragements PCRed on [[#11-05-2010|11-05-2010]], digested with BsaI-HF at 37<sup>o</sup>C for 1.5hours, heat inactivated at 65<sup>o</sup>C for 30 minutes. Tried to ligate KanA/B' fragments to each other and tried to ligate KanB/A' fragments to each other. Also tried to ligate KanA/B' fragments with KanB/A'. Ligated with T4 DNA ligase for 3 hours at 21<sup>o</sup>C. | ||
| + | |||
| + | Set up liquid cultures of KanRA/B'-Bsa and KanR B/A'-Bsa in pSB1C3 from plates streaked with on [[#30-05-2010|30-05-2010]] | ||
| + | |||
| + | ==02-06-2010== | ||
| + | |||
| + | Miniprepped liquid cultures from [[#01-06-2010|01-06-2010]]. Ran a 1% agarose gel of the ligations performed [[#01-06-2010|01-06-2010]]. | ||
| + | <!-- image of gel--> | ||
| + | To optimize the Restriction and ligation of BsaI-HF, digested KanA/B' and KanB/A' fragements PCRed on [[#11-05-2010|11-05-2010]] with the following recipe: | ||
| + | |||
| + | Digestion Recipe: | ||
| + | :14μL either A/B' or B/A' Kanamycin resistance cassette (approx. 100ng/μL) | ||
| + | :5μL 1/10 dillution of 100X BSA | ||
| + | :5μL 10X NEBuffer4 | ||
| + | :1.5&mu:L BsaI-HF | ||
| + | :24.5μL MilliQ | ||
| + | Digested at 50<sup>o</sup>C for 1hour, heat inactivated the enzyme at 65<sup>o</sup>C for 20 minutes | ||
| + | PCR purified the digests. | ||
| + | |||
| + | ==03-06-2010== | ||
| + | |||
| + | Tried to ligate the KanA/B' fragments to itself. Tried to ligate the KanB/A' fragments to itself. Tried to ligate the KanA/B' fragments to the KanB/A' fragments. | ||
| + | |||
| + | Ligation Recipe: | ||
| + | :8μL of digest from [[#02-06-2010|02-06-2010]] (either 8μL of one of the fragments or 4μL of each) | ||
| + | :1μL T4 DNA ligase | ||
| + | :6μL 5X Buffer | ||
| + | :15μL MilliQ H<sub>2</sub>O | ||
| + | |||
| + | Took aliquots of ligations at varying times and ran 1% agarose gels to test ligation. | ||
| + | <!--images--> | ||
| + | |||
| + | Digested some of Minipreps of the KanA/B' fragment inserted into pSB1C3 from [[#02-06-2010|02-06-2010]]. | ||
| + | Digestion Recipe: | ||
| + | :14μL plasmid (approx 300-400ng/&mu:L) | ||
| + | :5μL 10X NEBuffer 4 | ||
| + | :5μL 1/10 dilution of 100X BSA | ||
| + | :1.5μL BsaI-HF | ||
| + | :24.5μL MilliQ H<sub>2</sub>O | ||
| + | |||
| + | |||
| + | Made a 1/100 dilution of AmpR and TetR PCR Products from [[#27-05-2010|27-05-2010]] and performed PCR reactions to produce antibiotic inserts to make parts. | ||
| + | |||
| + | PCR Recipe: | ||
| + | :35.5μL MilliQ H<sub>2</sub>O | ||
| + | :1μL 10 uM dNTPs | ||
| + | :5μL 10X PCR Buffer | ||
| + | :2μL 50 uM MgCl<sub>2</sub> | ||
| + | :2.5μL Primer + (ApR 1/10+ or TR 1/10+) | ||
| + | :2.5μL Primer - (ApR 1/10- or TR 1/10-) | ||
| + | :1μL Template (1/100 diluted AmpR or TetR) | ||
| + | :0.5μL Taq Polymerase | ||
| + | |||
| + | PCR Program: | ||
| + | # 5 min-94<sup>o</sup>C | ||
| + | # 45 sec-94<sup>o</sup>C | ||
| + | # 1 min-60<sup>o</sup>C | ||
| + | # 1 min-72<sup>o</sup>C | ||
| + | # Repeat 2 through 4 35 times | ||
| + | # 5 min-72<sup>o</sup>C | ||
| + | |||
| + | ==04-06-2010== | ||
| + | |||
| + | Tried to test the limits of ligation reaction | ||
| + | Ligation Recipe of plasmids cut on [[#03-06-2010|03-06-2010]]: | ||
| + | :8μL digestion mixture | ||
| + | :6μL 5X T4 ligase buffer | ||
| + | :1μL T4 ligase | ||
| + | :15μL MilliQ | ||
| + | |||
| + | Tried to further reaction of KanA/B' fragments to KanB/A' fragments. To the existing Ligase reactions from [[#03-06-2010|03-06-2010]] added: | ||
| + | :1μL T4 ligase | ||
| + | :6μL 5X T4 ligase buffer | ||
| + | :23μL MilliQ | ||
| + | |||
| + | Tried to set limits of Kan fragments that would ligate. | ||
| + | :24μL digestion from [[#02-06-2010|02-06-2010]] (either 24μL of one of the fragments or 12μL of each) | ||
| + | :6μL 5X T4 ligase buffer | ||
| + | :1μL T4 ligase | ||
| + | |||
| + | <!--images--> | ||
| + | |||
| + | ==09-06-2010== | ||
| + | |||
| + | Restriction Digested AmpR and TetR inserts from [[#03-06-2010|03-06-2010]] to be ligated with psB1C3 vector later on. | ||
| + | |||
| + | Digestion Recipe for AmpR: | ||
| + | :13.4μL MilliQ H<sub>2</sub>O | ||
| + | :0.60&mu:L AmpR (333.3 ng/μL, determined by nanodrop) | ||
| + | :2μL 10X BSA | ||
| + | :2μL 10X Buffer 4 | ||
| + | :2μL BsaI | ||
| + | |||
| + | Digestion Recipe fro TetR: | ||
| + | :13.3μL MilliQ H<sub>2</sub>O | ||
| + | :0.7&mu:L TetR (571.3 ng/μL, determined by nanodrop) | ||
| + | :2μL 10X BSA | ||
| + | :2μL 10X Buffer 4 | ||
| + | :2μL BsaI | ||
| + | |||
| + | Both digestions were incubated at 50<sup>o</sup>C for 1hour, heat inactivated the enzyme at 70<sup>o</sup>C for 20 minutes. | ||
| + | |||
| + | ==10-06-2010== | ||
| + | |||
| + | Double Digested Kan/Chlor minipreps <!--Find date of Kan/Chlor production--> to determine orientation. | ||
| + | |||
| + | Digestion Recipe: | ||
| + | :5μL 10X Buffer 3 | ||
| + | :1μL XbaI | ||
| + | :1μL PstI | ||
| + | :5μL Kan/Chlor fragments | ||
| + | :33μL MilliQ H<sub>2</sub>O | ||
| + | :5μL 10X BSA | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1hour, heat inactivated the enzyme at 80<sup>o</sup>C for 20 minutes. | ||
| + | |||
| + | <---gel image of 10.06.10 Karina1---> | ||
| + | |||
| + | |||
| + | Restriction Digested A/B' psB1C3 vector. | ||
| + | |||
| + | Digestion Recipe: | ||
| + | :10μL MilliQ H<sub>2</sub>O | ||
| + | :5&mu:L psB1C3 | ||
| + | :2μL 10X BSA | ||
| + | :2μL 10X Buffer 4 | ||
| + | :1μL BsaHF | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1hour, heat inactivated the enzyme at 80<sup>o</sup>C for 20 minutes. | ||
| + | |||
| + | Ligated digested A/B' psB1C3 with AmpR from [[#09-06-2010|09-06-2010]]. | ||
| + | |||
| + | Ligation Recipe: | ||
| + | :9μL A/B' psB1C3 vector | ||
| + | :5μL AmpR insert | ||
| + | :6μL 5X Ligase Buffer | ||
| + | :1μL Ligase | ||
| + | :9μL MilliQ H<sub>2</sub>O | ||
| + | |||
| + | Incubated at room temperature for 45 minutes. Transformed with DH5α cells using 15μL of the ligated A/B' psB1C3 with AmpR. | ||
| + | |||
| + | ==11-06-2010== | ||
| + | |||
| + | We got colonies of psB1C3 with AmpR from the DH5α transformations from [[#10-06-2010|10-06-2010]]. Hooray~ We made overnights. | ||
| + | |||
| + | ==12-06-2010== | ||
| + | |||
| + | Miniprep of psB1C3 with AmpR was made from the overnights from [[#11-06-2010|11-06-2010]]. | ||
| + | |||
| + | ==16-06-2010== | ||
| + | |||
| + | psB1C3 with AmpR miniprep from [[#12-06-2010|12-06-2010]] ran undigested on a 1% agarose gel. | ||
| + | |||
| + | <!--gel image of Karina's 16.06.10--> | ||
| + | |||
| + | We digested psB1C3 with AmpR to see if the AmpR insert would be released from the psB1C3 vector. | ||
| + | |||
| + | Digestion Recipe: | ||
| + | :14.1μL MilliQ H<sub>2</sub>O | ||
| + | :0.9μL psB1C3 with AmpR (222.5 ng/μL, determined by nanodrop) | ||
| + | :2μL 10X BSA | ||
| + | :2μL 10X Buffer 4 | ||
| + | :1μL BsaI | ||
| + | |||
| + | Incubated at 50<sup>o</sup>C for 1hour, heat inactivated the enzyme at 65<sup>o</sup>C for 30 minutes. | ||
| + | |||
| + | Also, we made more overnights of psB1C3 with AmpR. | ||
| + | |||
| + | ==17-06-2010== | ||
| + | |||
| + | Digested psB1C3 with AmpR from [[#16-06-2010|16-06-2010]] was ran on a 1% agarose gel. | ||
| + | |||
| + | <!--17.06.10 Anh--> | ||
| + | |||
| + | ==28-06-2010== | ||
| + | <p>Transformed 2009 Cambridge color series parts: Bba_K274100, BBa_K274200, BBa_K274002, BBa_K274003, BBa_K274004, BBa_J23100. Also transformed 2004 UTAustin BBa_M30109. Transformed using 5uL of DNA.</p> | ||
| + | <p>Made starter culture of Dbl3 in LB at 11:03am. Innoculated large overnight liquid culture as per specifications in Inoue method at 6:00pm</p> | ||
| + | <p>Performed an enzyme efficiency experiment to determine which of BbsI, BfuAI, BsaI, or BsaI-HF works most efficiently. </p> | ||
| + | |||
| + | ==29-06-2010== | ||
| + | |||
| + | <p>Made a glycerol stock of PL5 from 1.5mL liquid overnight made [[#28-06-2010|28-06-2010]]. </p> | ||
| + | <p> Took remaining transformation of Dbl3 cells with ccdB BfuA/B' and ccdB BfuB/A' from [[#28-06-2010|28-06-2010]] out of the fridge and plated 25, 50 and 100μL of each on chloramphenicol LB plates</p> | ||
| + | <p>From the chloramphenicol and chloramphenicol/Kanamycin plates streaked with ccdB BbsA/B', ccdB BbsB/A',ccdB BsaA/B' and ccdB BsaB/A' on [[#28-06-2010|28-06-2010]], only ccdB BsaB/A' #6,8,10,11,12,14,15,18, ccdB BbsA/B' #8,15 and 20 grew on only the chloramphenicol plate. Made 5mL overnight liquid cultures with chloramphenicol of the some of the streaks that worked (ccdB BsaB/A' #6,8,10,11,12, ccdB BbsA/B' # 15, 20 and ccdB BsaB/A'#5 which didn't work but is a positive control).</p> | ||
| + | <p>Miniprepped liquid culture grown [[#28-06-2010|28-06-2010]] of Kan BsaA/B' #7. Concentration is 293.4ng/μL.</p> | ||
| + | <p>All the transormations of Cambridge 1009 color series performed [[#28-06-2010|28-06-2010]] worked. Made 5mL liquid cultures of each part.</p> | ||
| + | <p>Made competent Dbl3 cells by the Inuoe method from liquid cultures grown overnight. (set up on [[#28-06-2010|28-06-2010]]). The OD of the culture was 0.775.</p> | ||
| + | |||
| + | ==30-06-2010== | ||
| + | |||
| + | <p>Re Ran enzyme efficiency experiment with the same specifications as on [[#28-06-2010|28-06-2010]] but omitting BsaI-HF from the experiment because BsaI, BbsI and BfuAI appear to be the best of the enzymes. Also we ran the experiment with 10 Units of each enzyme not 20 Units.</p> | ||
| + | <p>Miniprepped liquid cultures of ccdB Bbs A/B' # 15 and 20 and of ccdB BsaB/A'#6, 8, 10, 11, 12 and 5(which did grow on both Kan/Chlor and Chlor plates, so it is a positive control for the incorrect plasmid) grown [[#29-06-2010|29-06-2010]]. The concentrations of the minipreps were between 200 and 400ng/mu;L and the curves did not show contamination. Digested approximately 500ng of minipreps with EcoRI and PstI with the following recipe: | ||
| + | :19.5μL MilliQ H<sub>2</sub>O | ||
| + | :2.5&mu:L Miniprep | ||
| + | :3μL 10X BSA | ||
| + | :3μL 10X NEBuffer 3 | ||
| + | :1μL EcoRI | ||
| + | :1μL PstI | ||
| + | </p> | ||
| + | <p>made 5mL culture of Kan BbsA/B' #1 from [[#]]</p> | ||
| + | <p>Ya! We got colonies of the ccdB BfuA/B' an ccdB BfuB/A' part plasmids that were plated [[#29-06-2010|29-06-2010]].</p> | ||
| + | <p>Miniprep of liquid cultures made [[#29-06-2010|29-06-2010]] of transformations of Cambridge 2009 color series. Concentrations are as follows: J23100, 397.7 ng/μL; K274100, 388.9ng/μL; K274200, 556.5ng/μL; K274004, 106.1ng/μL; K130109, 82.5ng/μL; K274003, 161.0ng/μL.</p> | ||
| + | |||
| + | ==01-07-2010== | ||
| + | |||
| + | Made 5mL liquid cultures with Choramphenicol with streaks from plates of ccdB Bbs A/B', ccdB Bbs B/A', cddB BsaA/B', ccdB BsaB/A' part plasmids from [[#25-06-2010|25-06-2010]]. Incubated at 37<sup>o</sup>C for 1 hour. | ||
| + | Plated 25μL of 1/100 and 1/1000 dilutions of liquid cultures onto Chloramphenicol LB plates. | ||
| + | |||
| + | Miniprepped liquid culture of Kan BbsA/B' #1 made [[#30-06-2010|30-06-2010]]. | ||
| + | |||
| + | Ran gel of Enzyme efficiency experiment performed on [[#30-06-2010|30-06-2010]]. <!--Image w/ explanation--> | ||
| + | |||
| + | Ran gel of digests of Minipreps of ccdB part plasmids performed on [[#30-06-2010|30-06-2010]]to ensure they are as expected. <!-- Image w/ explanation--> | ||
| + | |||
| + | <!--AMANDA miniprep of ccdB BfuA/B', ccdB BfuB/A' part plasmids, digest with NotI?--> | ||
| + | |||
| + | ==02-07-2010== | ||
| + | |||
| + | The colonies on the 1/100 and 1/1000 dilution plates of ccdB Bbs A/B', ccdB Bbs B/A', cddB BsaA/B', ccdB BsaB/A' part plasmids made on [[#01-07-2010|01-07-2010]], were spaced appropriately so as to be able to pick individual colonies. We picked as many colonies as possible and streaked on both LB plates with Kanamycin and Chloramphenicol resistance and LB plates with only Choramphenicol resistance. Again to determine if ccdB was infact inserted. | ||
| + | |||
| + | ==Kan, Chlor, and Tet Parts== | ||
| + | Strategy: to insert Kan into pSB1A3 after cutting both with NotI. Then, digest with BsaI to take Kan out and insert chlor and tet parts. | ||
| + | |||
| + | ==30-06-2010== | ||
| + | |||
| + | =====Restriction Digest with NotI Protocol:===== | ||
| + | |||
| + | :Kan AB PCR product (82.3 ng/ul) 1ul | ||
| + | :pSB1A3 plasmid (30.5 ng/ul) 3ul | ||
| + | :NotI enzyme 1ul | ||
| + | :10x REact3 buffer 1ul | ||
| + | :MilliQ H2O 4ul | ||
| + | :Total 10ul | ||
| + | :Incubated at 37C for 1 1/2 hours. | ||
| + | |||
| + | =====Ligation Protocol:===== | ||
| + | |||
| + | :Digested pSB1A3 and Kan AB fragment 8ul | ||
| + | :T4 DNA ligase 1ul | ||
| + | :5X T4 DNA ligase buffer 6ul | ||
| + | :MilliQ H20 5ul | ||
| + | :Total 20ul | ||
| + | :Incubated at room temperature for 1 hour. | ||
| + | |||
| + | =====Gel Electrophoresis:===== | ||
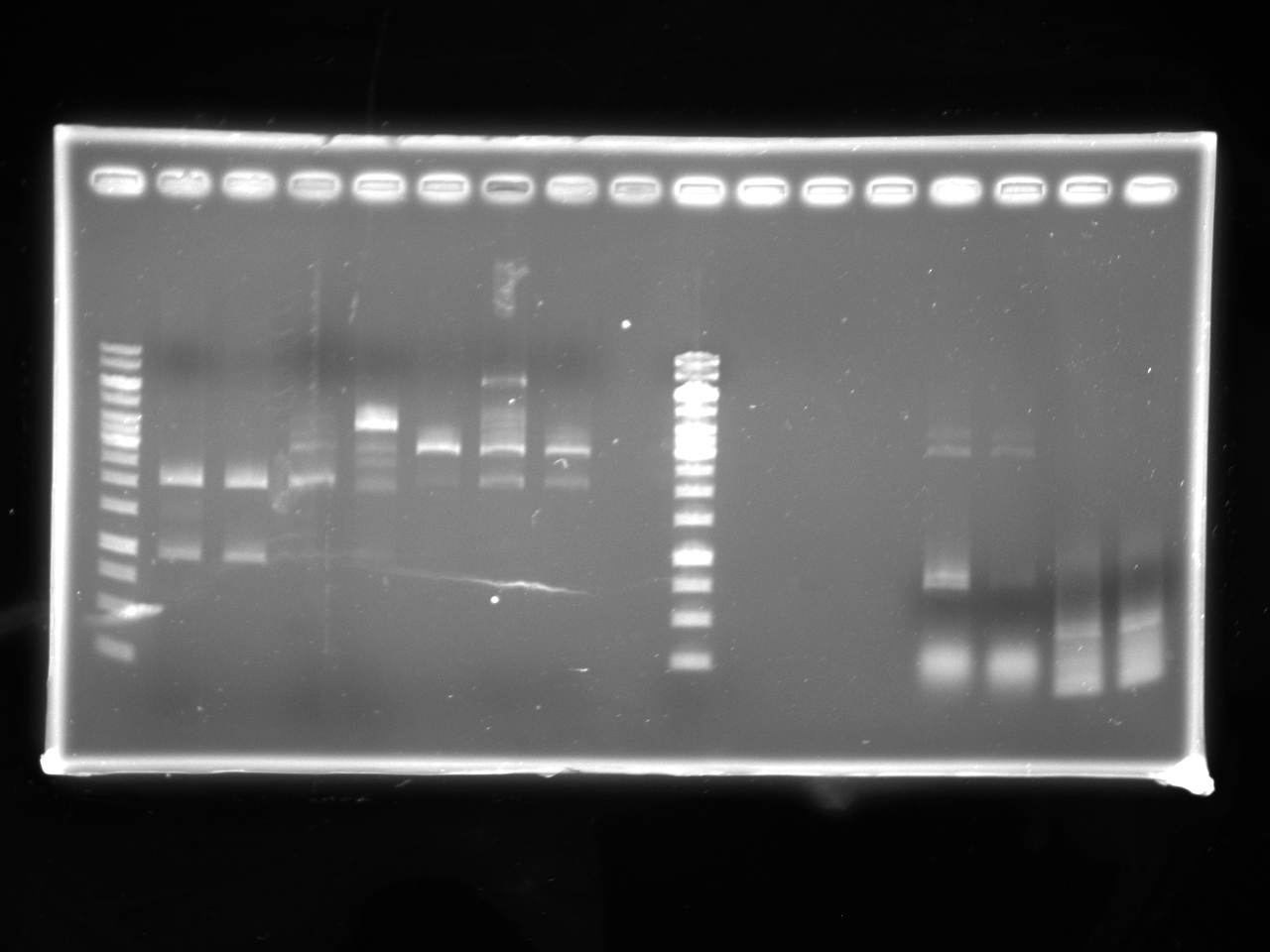
| + | [[Image:Uofa_igem_29.06.10.alina2.jpg|thumb|none|300px|Lanes 2-4 are digests of pSB1A3 and kan AB fragment with NotI. Lanes 5-7 are ligations of the digested pSB1A3 and kan fragments. Note that no ligated DNA shows up on the gel due to a small volume of digestion (3ul) loaded.]] | ||
| + | |||
| + | =====Transformation:===== | ||
| + | :Followed Inoue Protocol using 50ul DH5-alpha competent cells and 2ul of ligated pSB1A3 and kan AB part. | ||
| + | :Plated 100ul, 100ul, and 400ul on Kan/Amp plates overnight at 37C. | ||
| + | |||
| + | :Results: | ||
| + | ::Plate 1 (100ul): TNTC white colonies and ~20 red colonies | ||
| + | ::Plate 2 (100ul): TNTC white colonies and ~3 red colonies | ||
| + | ::Plate 3 (400ul): TNTC white colonies and ~30 red colonies | ||
| + | |||
| + | ==06-07-2010== | ||
| + | |||
| + | =====Overnights:===== | ||
| + | :Set up 5ml overnight tubes with 5ml LB broth, 5ul kanamycin, 2.5ul ampicillin. | ||
| + | |||
| + | :Results: | ||
| + | ::All seven tubes had growth and no growth in control tube. | ||
| + | |||
| + | |||
| + | ==07-07-2010== | ||
| + | |||
| + | =====Minipreps===== | ||
| + | Followed Fermentas protocol to prepare 7 tubes. | ||
| + | |||
| + | =====Restriction Digest with XbaI and PstI Protocol to check for orientation:===== | ||
| + | |||
| + | :Kan AB in pSB1A3 1ul | ||
| + | :XbaI enzyme 2ul | ||
| + | :PstI enzyme 1ul | ||
| + | :NEBuffer 3 4ul | ||
| + | :MilliQ H2O 3ul | ||
| + | :BSA 4ul | ||
| + | :Total 14ul | ||
| + | :Incubated at 37C for 1 1/2 hours. | ||
| + | |||
| + | =====Gel Electrophoresis===== | ||
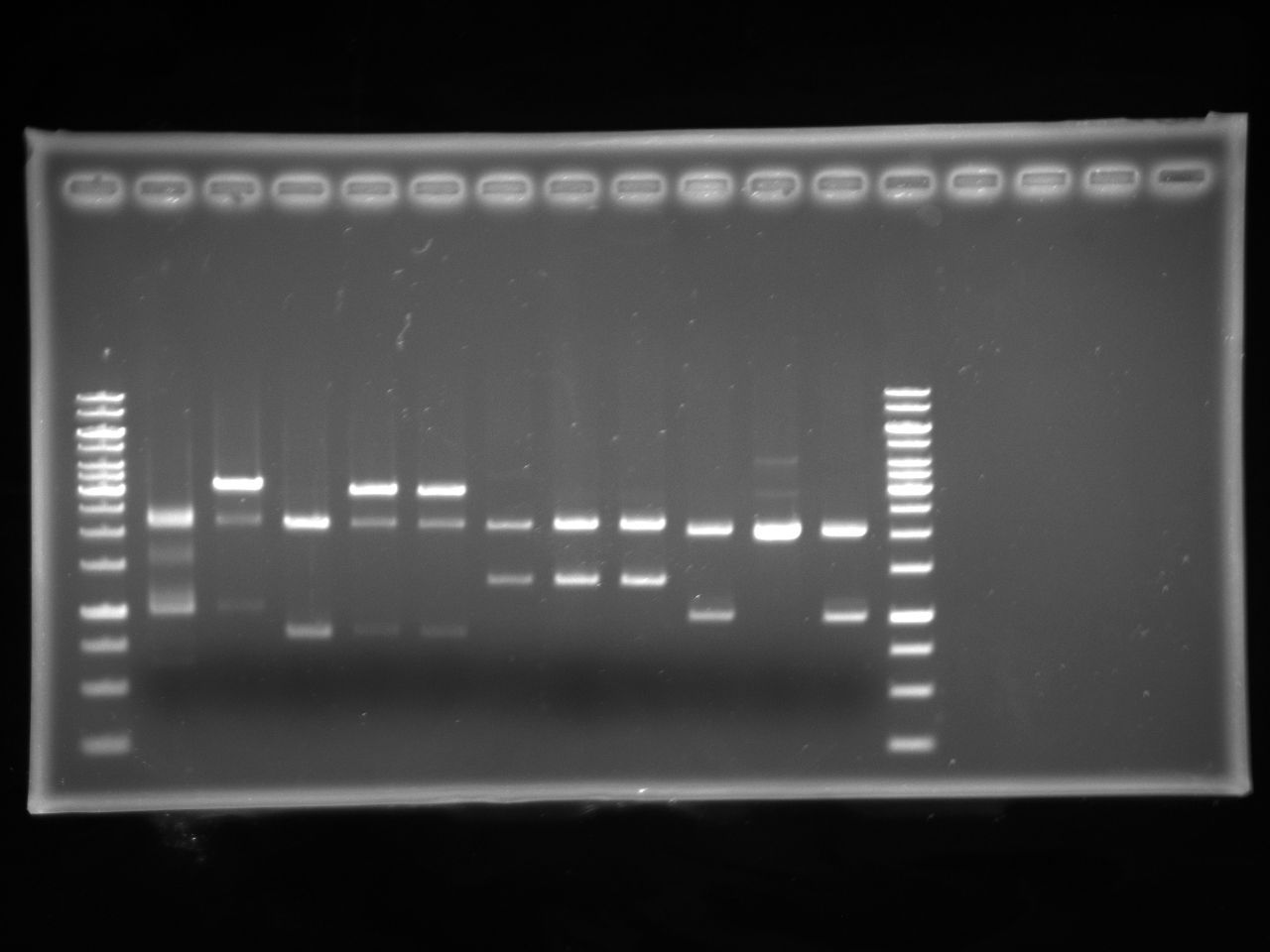
| + | [[Image:Uofa_iGem_08.07,10alinakarina.jpg|thumb|none|300px|This is a shared gel. Lanes 2-8 are pSB1A3 plasmids with AB Kan inserts from different tubes, digested with XbaI and PstI. The plasmids from lanes 2 and 3 produced proper fragments. The rest of the tubes were discarded.]] | ||
| + | |||
| + | =====Restriction Digest with BsaI Protocol:===== | ||
| + | |||
| + | :Kan AB in pSB1A3 (247.4 ng/ul or 255.8 ng/ul) 1ul | ||
| + | :chlor or tet AB fragment 1ul | ||
| + | :BsaI enzyme 0.5ul | ||
| + | :NEBuffer 4 3ul | ||
| + | :MilliQ H2O 5.5ul | ||
| + | :Total 11ul | ||
| + | :Incubated at 50C for 1 hour. | ||
| + | |||
| + | =====Ligation Protocol:===== | ||
| + | |||
| + | :Digested pSB1A3 with Kan AB and chlor or tet 8ul | ||
| + | :T4 DNA ligase 1ul | ||
| + | :5X T4 DNA ligase buffer 6ul | ||
| + | :MilliQ H20 5ul | ||
| + | :Total 20ul | ||
| + | :Incubated at room temperature for 1 hour. | ||
| + | |||
| + | =====Gel Electrophoresis===== | ||
| + | Insert gel here. | ||
| + | |||
| + | ==08-07-2010== | ||
| + | |||
| + | =====Transformations===== | ||
| + | :Followed Inoue transformation protocol using 50ul DH5-alpha competent cells and 2ul of pSB1A3 ligated with tet or chlor. | ||
| + | :Plated 200ul and 600ul on chlor/amp plates for chlor insert and on tet/amp plates for tet insert. | ||
| + | |||
| + | :Results: | ||
| + | ::chlor/amp plate 1 (200ul): ~300 colonies | ||
| + | ::chlor/amp plate 2 (600ul): TNTC colonies | ||
| + | ::tet/amp plate 1 (200ul): ~20 colonies | ||
| + | ::tet/amp plate 2 (600ul): ~30 colonies | ||
| + | |||
| + | ==09-07-2010== | ||
| + | |||
| + | =====Overnights===== | ||
| + | :5ml LB, [20ul chlor and 2.5 ul amp] or [20ul tet and 2.5 ul amp] | ||
| + | |||
| + | ::Results | ||
| + | :growth in all experimental tubes and no growth in control tubes | ||
| + | |||
| + | ==10-07-2010== | ||
| + | |||
| + | =====Minipreps===== | ||
| + | 4 tubes of tet in pSB1A3 were prepared and 7 tubes of chlor in pSB1A3 were prepared. | ||
| + | |||
| + | ==12-07-2010== | ||
| + | |||
| + | =====Restriction Digest with EcoRI Protocol to determine plasmid size:===== | ||
| + | |||
| + | :pSB1A3 with chlor or tet insert 1ul | ||
| + | :Fast Digest EcoRI 1ul | ||
| + | :10X Fast Digest Buffer 2ul | ||
| + | :MilliQ H2O 16ul | ||
| + | :Total 20ul | ||
| + | :Incubated at 37C for 5 mins. | ||
| + | |||
| + | =====Gel Electrophoresis===== | ||
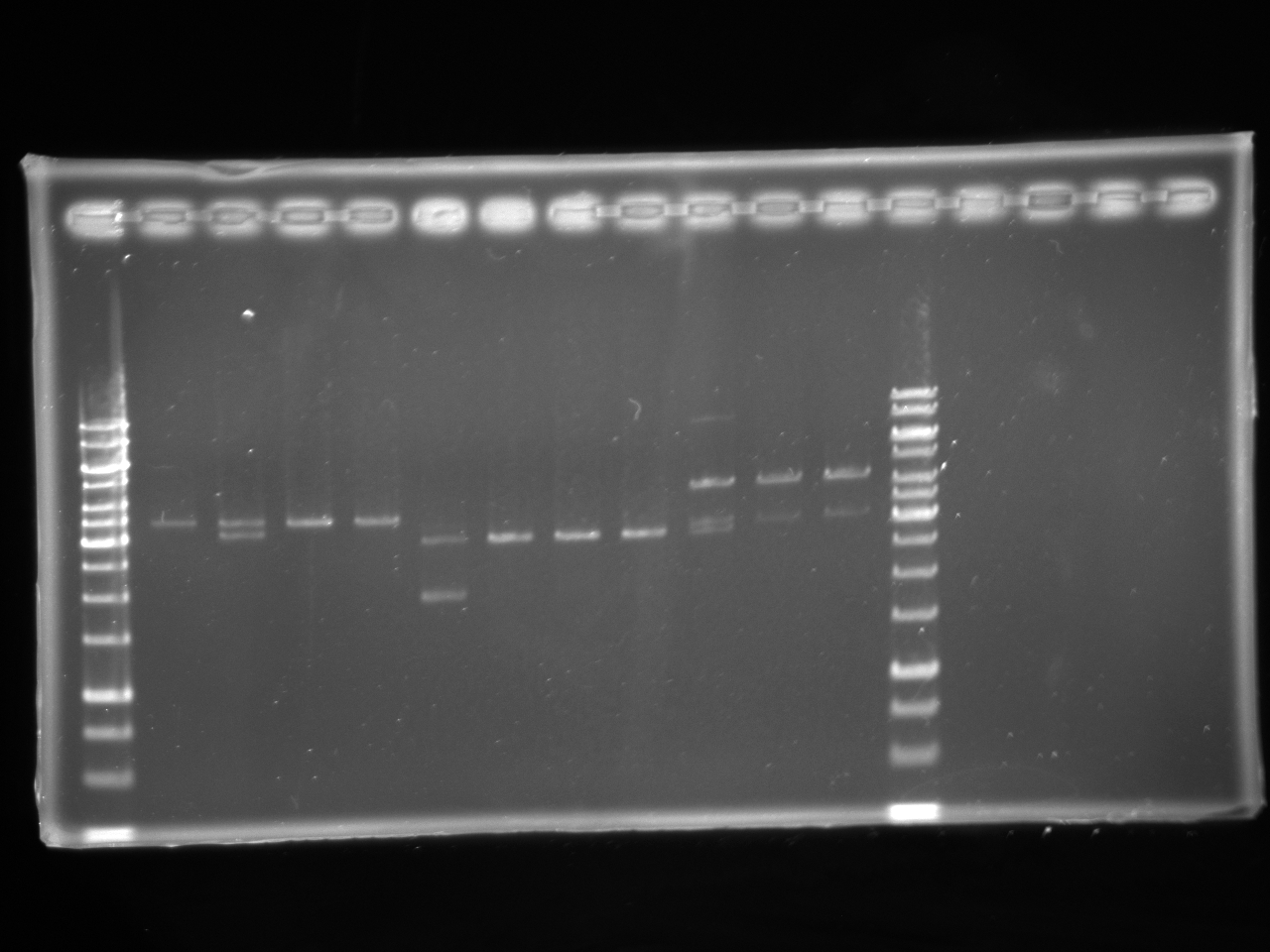
| + | [[Image:Uofa_iGEM_12.07.10.alina.jpg|thumb|none|300px|Lanes 2-5 are pSB1A3 plasmid with tet AB insert. Lanes 6-13 are pSB1A3 plasmid with chlor AB insert. All plasmids were digested with EcoRI. All but the products of lanes 3,6,10,11,12 were correct. 1 out of the 4 tet in pSB1A3 tubes cut incorrectly and was discarded, 4 out of the 7 chlor in pSB1A3 tubes cut incorrectly and was discarded.]] | ||
| + | |||
| + | ==14-07-2010== | ||
| + | |||
| + | =====Double Digest of chlor, kan and tet in PSB1A3 and amp is pSB1C3 with Xba and PstI protocol===== | ||
| + | |||
| + | :plasmid with antibiotic part 1ul | ||
| + | :10X NEBuffer 3 1ul | ||
| + | :MilliQ H20 5ul | ||
| + | :XbaI 1ul | ||
| + | :PstI 1ul | ||
| + | :10X BSA 1ul | ||
| + | :Total 10ul | ||
| + | :Incubated at 37C for 1 hour. | ||
| + | |||
| + | =====Gel Electrophoresis===== | ||
| + | [[Image:Uofa igem 14.07.10.alina.jpg|thumb|none|300px|Digests of antibiotic AB fragments with XbaI and PstI. Lanes 2-3 are kanamycin AB, lanes 4-6 are chloramphenicol AB, lanes 7-9 are tetracycline AB, lanes 10-12 are ampicillin AB. All of the plasmids cut correctly except for one of the amp in pSB1C3, which was discarded]] | ||
| + | |||
| + | ==15-07-2010== | ||
| + | :Antibiotic parts are now stored in the DNA box according to the BioBytes registry. | ||
| + | :Glycerol stocks were made of overnights from chlor, tet, and kan in PSB1A3 plates. | ||
| + | |||
| + | |||
| + | {| style="color:#1b2c8a;background-color:#FFFF33;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Team:Alberta/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Alberta/Building Parts|Building Parts]] | ||
| + | !align="center"|[[Team:Alberta/Testing Parts|Testing Parts]] | ||
| + | !align="center"|[[Team:Alberta/Assembly Method|Assembly Method]] | ||
| + | !align="center"|[[Team:Alberta/Competent Cells|Competent Cells]] | ||
| + | !align="center"|[[Team:Alberta/Plates|Plates]] | ||
| + | !align="center"|[[Team:Alberta/Kit Manual|Kit Manual]] | ||
| + | !align="center"|[[Team:Alberta/Software|Software]] | ||
| + | |} | ||
Latest revision as of 19:55, 19 August 2010

Our original base plasmids contain a Kanamycin resistance cassette (p1003) bracketed by BsaI or BfuAI or BbsI cut sites. If cut with either BsaI or BfuAI or BbsI, the Kanamycin cassette is release with sticky ends characteristic of an A or a B BioByte.
May 10, 2010 PCRed p1003 (Kanamycin cassette)with primers PrA_p1003+ and PrB'_p1003-
May 10, 2010 PCRed p1003 (Kanamycin cassette)with primers PrB_p1003+ and PrA'_p1003-
May 19, 2010 Digested pSB1C3 with Not1
May 19, 2010 Digested PCR products of p1003 (Kanamycin cassette) Not1
May 25, 2010 Ligated PCR products of p1003 (Kanamycin cassette) and pSB1C3
Before we were able to test parts we created 2 base testing plasmids (vector 01 and vector 02). Vector 01 is designed to test Open Reading Frame parts, or parts that code for proteins. The part is flanked by a promoter and the start codon on one side and a stop codon and terminator on the other. Vector 02 is designed to test linker parts, or parts that control the expression of the Open Reading Frame parts they are next two. In Vector 02 the part is flanked by two distinct reporter genes, that by comparing the relative expression of the 2 reporter genes we can determine the behavior of the linking part.
 Oscar Cortes
Oscar CortesSpecialized in Molecular Genetics (Graduate)
Building Parts
10-05-2010
PCR a Kanamycin resistance cassette fragment from p1003 with primers containing either the A/B' ends or the B/A' ends. (Fragments formed called KanR A/B'-Bsa and KanR B/A'-Bsa respectively)
Recipe:
- 1μL p1003 (approx. 1ng)
- 2.5μL prA_p1003+
- 2.5μL prB'_p1003-
- 5μL 10X PCR buffer
- 1μL 10uM dNTPs
- 2μL 50uM MgCl2
- 0.5μL Taq polymerase
- 35.5μL MilliQ H2O
Same recipe for KanR B/A'-Bsa except primers are prB_p1003+ and prA'_p1003-.
Program:
- 5 min-94oC
- 45 sec-94oC
- 1 min-60oC
- 1 min-72oC
- Repeat 2 through 4 35 times
- 5 min-72oC
11-05-2010
PCR purification of KanR A/B'-Bsa and KanR B/A'-Bsa created 10-05-2010 with Qiagen PCR cleanup kit.
Determined concentrations by nanodrop. KanA/B'-Bsa: 101.1ng/μL KanB/A'-Bsa:89.6ng/μL
17-05-2010
Innoculated 250mL overnight cultures with 10mL,4mL and 2mL of a starter culture of DH5α. Left shaking at 18oC overnight.
18-05-2010
Prepared DH5α E.Coli competent cells using the Inoue Method.
Transformed DH5α cells with pSB1C3-J04450 and grew overnight at 37oC on Chloramphenicol plates
19-05-2010
From the transformation of DH5α cells with pSB1C3-J04450 performed on 18-05-2010, we took 4 distinct colonies, streaked them on a new chloramphenicol plate and inoculated 5ml liquid cultures.
20-05-2010
Performed a Miniprep of 3 of the 4 5ml liquid cultures of DH5α cells with pSB1C3-J04450 from 19-05-2010. Took a 1μL sample of the Miniprep solutions and digested with NotI at 37oC for 1 hour.
Digestion Recipe:
- 1μL Miniprep (between 153.2 ng/μl and 302.7ng/μl determined by nanodrop)
- 1μL NotI
- 1μL 10X ReACT 3
- 7μL MilliQ
Ran Digestion on a 1% agarose gel to check that the plasmid obtained with what we expected.
25-05-2010
Made 1.5mL LB liquid cultures of pSB1C3 from the plate streaked on 19-05-2010 and added chloramphenicol.
26-05-2010
Made 3 glycerol stocks of pSB1C3 from overnight made 25-05-2010
27-05-2010
</p>Digested both A/B' and B/A' Kanamycin Resistance cassettes fragments from 11-05-2010 and pSB1C3 from 20-05-2010 with NotI at 37oC for 1 hour. Heat inactivated the NotI for 10 minutes at 65oC. Ligated the Kanamycin Resistance cassettes into pSB1C3 at 16oC for 1 hour then took 15μL to room temperature for 2 hours. Transformed 100μL of DH5α cells with 5μL of RT ligation reaction. Plated transformation on plates with both Chloramphenicol and Kanamycin.</p>
Digestion Recipe:
- 1μL Miniprep (302.7ng/μl determined by nanodrop)
- 2μL either A/B' or B/A' Kanamycin resistance cassette (approx. 100ng/μL)
- 1μL NotI
- 1μL 10X ReACT 3
- 5μL MilliQ
Ligation Recipe:
- 10μL of Digest solution
- 1μL T4 DNA ligase
- 6μL 5X Buffer
- 13μL MilliQ H2O
Also transformed pSB4A5-J04450, pSB4C5-J04450 and pSB3T5-J04450 from the 2010 biobrick parts into DH5α cells.
Performed PCR reactions to create parts with antibiotic resistance with negative controls.
PCR Recipe:
- 3μL 10X PCR Buffer
- 1μL 10 uM dNTPs
- 2μL 50 uM MgCl2
- 17.5μL MilliQ H2O
- 0.5μL Taq Polymerase
- 1μL Template (psB4A5-J04450, psB4C5-J04450 or psB3T5-J04450)
- 2.5μL Primer + (PrA psB4A5 ApR+, PrA psB4C5 ChR+ or PrA psB3T5 TR+)
- 2.5μL Primer - (PrB psB4A5 ApR-, PrB psB4C5 ChR- or PrB psB3T5 TR-)
PCR Program:
- 5 min-94oC
- 45 sec-94oC
- 1 min-60oC
- 1 min-72oC
- Repeat 2 through 4 35 times
- 5 min-72oC
28-05-2010
We got colonies!! (it's a fantastic feeling) We then lovingly put them in the cold room to await our return from Calgary
30-05-2010
From the transformation of DH5α cells with pSB1C3-KanR performed on 28-05-2010, we took 12 distinct colonies of each KanA/B' and KanB/A', streaked them on a new chloramphenicol plate and inoculated 5ml liquid cultures with the appropriate antibiotics overnight at 37oC. We also picked colonies of pSB4A5-J04450, pSB4C5-J04450 and pSB3T5-J04450, streaked and made 5mL liquid cultures of them too.
31-05-2010
9/12 of the pSB1C3-KanA/B' Liquid cultures 30-05-2010 were successful and only 1/12 of the pSB1C3-KanB/A' liquid cultures were successful. The pSB4A5, pSB3T5 and pSB4C5 liquid cultures worked. Miniprepped all the liquid cultures that worked. However, the streaks on the plates worked.
Performed a restriction digest of an aliquot of the pSB1C3-KanA/B' and pSB1C3-KanB/A' minipreps. Digested with XbaI at 37oC for one hour and then with EcoRI at 37oC for one hour. Ran a 1% agarose gel of the digests to determine the orientation of the KanR fragments.
KanA/B' and KanB/A' fragements PCRed on 11-05-2010, digested with BsaI at 37oC for 1.5hours, heat inactivated at 65oC for 30 minutes. Tried to ligate KanA/B' fragments to each other and tried to ligate KanB/A' fragments to each other. Also tried to ligate KanA/B' fragments with KanB/A'. Ligated with T4 DNA ligase overnight at 16oC.
01-06-2010
KanA/B' and KanB/A' fragements PCRed on 11-05-2010, digested with BsaI-HF at 37oC for 1.5hours, heat inactivated at 65oC for 30 minutes. Tried to ligate KanA/B' fragments to each other and tried to ligate KanB/A' fragments to each other. Also tried to ligate KanA/B' fragments with KanB/A'. Ligated with T4 DNA ligase for 3 hours at 21oC.
Set up liquid cultures of KanRA/B'-Bsa and KanR B/A'-Bsa in pSB1C3 from plates streaked with on 30-05-2010
02-06-2010
Miniprepped liquid cultures from 01-06-2010. Ran a 1% agarose gel of the ligations performed 01-06-2010. To optimize the Restriction and ligation of BsaI-HF, digested KanA/B' and KanB/A' fragements PCRed on 11-05-2010 with the following recipe:
Digestion Recipe:
- 14μL either A/B' or B/A' Kanamycin resistance cassette (approx. 100ng/μL)
- 5μL 1/10 dillution of 100X BSA
- 5μL 10X NEBuffer4
- 1.5&mu:L BsaI-HF
- 24.5μL MilliQ
Digested at 50oC for 1hour, heat inactivated the enzyme at 65oC for 20 minutes PCR purified the digests.
03-06-2010
Tried to ligate the KanA/B' fragments to itself. Tried to ligate the KanB/A' fragments to itself. Tried to ligate the KanA/B' fragments to the KanB/A' fragments.
Ligation Recipe:
- 8μL of digest from 02-06-2010 (either 8μL of one of the fragments or 4μL of each)
- 1μL T4 DNA ligase
- 6μL 5X Buffer
- 15μL MilliQ H2O
Took aliquots of ligations at varying times and ran 1% agarose gels to test ligation.
Digested some of Minipreps of the KanA/B' fragment inserted into pSB1C3 from 02-06-2010. Digestion Recipe:
- 14μL plasmid (approx 300-400ng/&mu:L)
- 5μL 10X NEBuffer 4
- 5μL 1/10 dilution of 100X BSA
- 1.5μL BsaI-HF
- 24.5μL MilliQ H2O
Made a 1/100 dilution of AmpR and TetR PCR Products from 27-05-2010 and performed PCR reactions to produce antibiotic inserts to make parts.
PCR Recipe:
- 35.5μL MilliQ H2O
- 1μL 10 uM dNTPs
- 5μL 10X PCR Buffer
- 2μL 50 uM MgCl2
- 2.5μL Primer + (ApR 1/10+ or TR 1/10+)
- 2.5μL Primer - (ApR 1/10- or TR 1/10-)
- 1μL Template (1/100 diluted AmpR or TetR)
- 0.5μL Taq Polymerase
PCR Program:
- 5 min-94oC
- 45 sec-94oC
- 1 min-60oC
- 1 min-72oC
- Repeat 2 through 4 35 times
- 5 min-72oC
04-06-2010
Tried to test the limits of ligation reaction Ligation Recipe of plasmids cut on 03-06-2010:
- 8μL digestion mixture
- 6μL 5X T4 ligase buffer
- 1μL T4 ligase
- 15μL MilliQ
Tried to further reaction of KanA/B' fragments to KanB/A' fragments. To the existing Ligase reactions from 03-06-2010 added:
- 1μL T4 ligase
- 6μL 5X T4 ligase buffer
- 23μL MilliQ
Tried to set limits of Kan fragments that would ligate.
- 24μL digestion from 02-06-2010 (either 24μL of one of the fragments or 12μL of each)
- 6μL 5X T4 ligase buffer
- 1μL T4 ligase
09-06-2010
Restriction Digested AmpR and TetR inserts from 03-06-2010 to be ligated with psB1C3 vector later on.
Digestion Recipe for AmpR:
- 13.4μL MilliQ H2O
- 0.60&mu:L AmpR (333.3 ng/μL, determined by nanodrop)
- 2μL 10X BSA
- 2μL 10X Buffer 4
- 2μL BsaI
Digestion Recipe fro TetR:
- 13.3μL MilliQ H2O
- 0.7&mu:L TetR (571.3 ng/μL, determined by nanodrop)
- 2μL 10X BSA
- 2μL 10X Buffer 4
- 2μL BsaI
Both digestions were incubated at 50oC for 1hour, heat inactivated the enzyme at 70oC for 20 minutes.
10-06-2010
Double Digested Kan/Chlor minipreps to determine orientation.
Digestion Recipe:
- 5μL 10X Buffer 3
- 1μL XbaI
- 1μL PstI
- 5μL Kan/Chlor fragments
- 33μL MilliQ H2O
- 5μL 10X BSA
Incubated at 37oC for 1hour, heat inactivated the enzyme at 80oC for 20 minutes.
<---gel image of 10.06.10 Karina1--->
Restriction Digested A/B' psB1C3 vector.
Digestion Recipe:
- 10μL MilliQ H2O
- 5&mu:L psB1C3
- 2μL 10X BSA
- 2μL 10X Buffer 4
- 1μL BsaHF
Incubated at 37oC for 1hour, heat inactivated the enzyme at 80oC for 20 minutes.
Ligated digested A/B' psB1C3 with AmpR from 09-06-2010.
Ligation Recipe:
- 9μL A/B' psB1C3 vector
- 5μL AmpR insert
- 6μL 5X Ligase Buffer
- 1μL Ligase
- 9μL MilliQ H2O
Incubated at room temperature for 45 minutes. Transformed with DH5α cells using 15μL of the ligated A/B' psB1C3 with AmpR.
11-06-2010
We got colonies of psB1C3 with AmpR from the DH5α transformations from 10-06-2010. Hooray~ We made overnights.
12-06-2010
Miniprep of psB1C3 with AmpR was made from the overnights from 11-06-2010.
16-06-2010
psB1C3 with AmpR miniprep from 12-06-2010 ran undigested on a 1% agarose gel.
We digested psB1C3 with AmpR to see if the AmpR insert would be released from the psB1C3 vector.
Digestion Recipe:
- 14.1μL MilliQ H2O
- 0.9μL psB1C3 with AmpR (222.5 ng/μL, determined by nanodrop)
- 2μL 10X BSA
- 2μL 10X Buffer 4
- 1μL BsaI
Incubated at 50oC for 1hour, heat inactivated the enzyme at 65oC for 30 minutes.
Also, we made more overnights of psB1C3 with AmpR.
17-06-2010
Digested psB1C3 with AmpR from 16-06-2010 was ran on a 1% agarose gel.
28-06-2010
Transformed 2009 Cambridge color series parts: Bba_K274100, BBa_K274200, BBa_K274002, BBa_K274003, BBa_K274004, BBa_J23100. Also transformed 2004 UTAustin BBa_M30109. Transformed using 5uL of DNA.
Made starter culture of Dbl3 in LB at 11:03am. Innoculated large overnight liquid culture as per specifications in Inoue method at 6:00pm
Performed an enzyme efficiency experiment to determine which of BbsI, BfuAI, BsaI, or BsaI-HF works most efficiently.
29-06-2010
Made a glycerol stock of PL5 from 1.5mL liquid overnight made 28-06-2010.
Took remaining transformation of Dbl3 cells with ccdB BfuA/B' and ccdB BfuB/A' from 28-06-2010 out of the fridge and plated 25, 50 and 100μL of each on chloramphenicol LB plates
From the chloramphenicol and chloramphenicol/Kanamycin plates streaked with ccdB BbsA/B', ccdB BbsB/A',ccdB BsaA/B' and ccdB BsaB/A' on 28-06-2010, only ccdB BsaB/A' #6,8,10,11,12,14,15,18, ccdB BbsA/B' #8,15 and 20 grew on only the chloramphenicol plate. Made 5mL overnight liquid cultures with chloramphenicol of the some of the streaks that worked (ccdB BsaB/A' #6,8,10,11,12, ccdB BbsA/B' # 15, 20 and ccdB BsaB/A'#5 which didn't work but is a positive control).
Miniprepped liquid culture grown 28-06-2010 of Kan BsaA/B' #7. Concentration is 293.4ng/μL.
All the transormations of Cambridge 1009 color series performed 28-06-2010 worked. Made 5mL liquid cultures of each part.
Made competent Dbl3 cells by the Inuoe method from liquid cultures grown overnight. (set up on 28-06-2010). The OD of the culture was 0.775.
30-06-2010
Re Ran enzyme efficiency experiment with the same specifications as on 28-06-2010 but omitting BsaI-HF from the experiment because BsaI, BbsI and BfuAI appear to be the best of the enzymes. Also we ran the experiment with 10 Units of each enzyme not 20 Units.
Miniprepped liquid cultures of ccdB Bbs A/B' # 15 and 20 and of ccdB BsaB/A'#6, 8, 10, 11, 12 and 5(which did grow on both Kan/Chlor and Chlor plates, so it is a positive control for the incorrect plasmid) grown 29-06-2010. The concentrations of the minipreps were between 200 and 400ng/mu;L and the curves did not show contamination. Digested approximately 500ng of minipreps with EcoRI and PstI with the following recipe:
- 19.5μL MilliQ H2O
- 2.5&mu:L Miniprep
- 3μL 10X BSA
- 3μL 10X NEBuffer 3
- 1μL EcoRI
- 1μL PstI
made 5mL culture of Kan BbsA/B' #1 from #
Ya! We got colonies of the ccdB BfuA/B' an ccdB BfuB/A' part plasmids that were plated 29-06-2010.
Miniprep of liquid cultures made 29-06-2010 of transformations of Cambridge 2009 color series. Concentrations are as follows: J23100, 397.7 ng/μL; K274100, 388.9ng/μL; K274200, 556.5ng/μL; K274004, 106.1ng/μL; K130109, 82.5ng/μL; K274003, 161.0ng/μL.
01-07-2010
Made 5mL liquid cultures with Choramphenicol with streaks from plates of ccdB Bbs A/B', ccdB Bbs B/A', cddB BsaA/B', ccdB BsaB/A' part plasmids from 25-06-2010. Incubated at 37oC for 1 hour. Plated 25μL of 1/100 and 1/1000 dilutions of liquid cultures onto Chloramphenicol LB plates.
Miniprepped liquid culture of Kan BbsA/B' #1 made 30-06-2010.
Ran gel of Enzyme efficiency experiment performed on 30-06-2010.
Ran gel of digests of Minipreps of ccdB part plasmids performed on 30-06-2010to ensure they are as expected.
02-07-2010
The colonies on the 1/100 and 1/1000 dilution plates of ccdB Bbs A/B', ccdB Bbs B/A', cddB BsaA/B', ccdB BsaB/A' part plasmids made on 01-07-2010, were spaced appropriately so as to be able to pick individual colonies. We picked as many colonies as possible and streaked on both LB plates with Kanamycin and Chloramphenicol resistance and LB plates with only Choramphenicol resistance. Again to determine if ccdB was infact inserted.
Kan, Chlor, and Tet Parts
Strategy: to insert Kan into pSB1A3 after cutting both with NotI. Then, digest with BsaI to take Kan out and insert chlor and tet parts.
30-06-2010
Restriction Digest with NotI Protocol:
- Kan AB PCR product (82.3 ng/ul) 1ul
- pSB1A3 plasmid (30.5 ng/ul) 3ul
- NotI enzyme 1ul
- 10x REact3 buffer 1ul
- MilliQ H2O 4ul
- Total 10ul
- Incubated at 37C for 1 1/2 hours.
Ligation Protocol:
- Digested pSB1A3 and Kan AB fragment 8ul
- T4 DNA ligase 1ul
- 5X T4 DNA ligase buffer 6ul
- MilliQ H20 5ul
- Total 20ul
- Incubated at room temperature for 1 hour.
Gel Electrophoresis:
Transformation:
- Followed Inoue Protocol using 50ul DH5-alpha competent cells and 2ul of ligated pSB1A3 and kan AB part.
- Plated 100ul, 100ul, and 400ul on Kan/Amp plates overnight at 37C.
- Results:
- Plate 1 (100ul): TNTC white colonies and ~20 red colonies
- Plate 2 (100ul): TNTC white colonies and ~3 red colonies
- Plate 3 (400ul): TNTC white colonies and ~30 red colonies
06-07-2010
Overnights:
- Set up 5ml overnight tubes with 5ml LB broth, 5ul kanamycin, 2.5ul ampicillin.
- Results:
- All seven tubes had growth and no growth in control tube.
07-07-2010
Minipreps
Followed Fermentas protocol to prepare 7 tubes.
Restriction Digest with XbaI and PstI Protocol to check for orientation:
- Kan AB in pSB1A3 1ul
- XbaI enzyme 2ul
- PstI enzyme 1ul
- NEBuffer 3 4ul
- MilliQ H2O 3ul
- BSA 4ul
- Total 14ul
- Incubated at 37C for 1 1/2 hours.
Gel Electrophoresis
Restriction Digest with BsaI Protocol:
- Kan AB in pSB1A3 (247.4 ng/ul or 255.8 ng/ul) 1ul
- chlor or tet AB fragment 1ul
- BsaI enzyme 0.5ul
- NEBuffer 4 3ul
- MilliQ H2O 5.5ul
- Total 11ul
- Incubated at 50C for 1 hour.
Ligation Protocol:
- Digested pSB1A3 with Kan AB and chlor or tet 8ul
- T4 DNA ligase 1ul
- 5X T4 DNA ligase buffer 6ul
- MilliQ H20 5ul
- Total 20ul
- Incubated at room temperature for 1 hour.
Gel Electrophoresis
Insert gel here.
08-07-2010
Transformations
- Followed Inoue transformation protocol using 50ul DH5-alpha competent cells and 2ul of pSB1A3 ligated with tet or chlor.
- Plated 200ul and 600ul on chlor/amp plates for chlor insert and on tet/amp plates for tet insert.
- Results:
- chlor/amp plate 1 (200ul): ~300 colonies
- chlor/amp plate 2 (600ul): TNTC colonies
- tet/amp plate 1 (200ul): ~20 colonies
- tet/amp plate 2 (600ul): ~30 colonies
09-07-2010
Overnights
- 5ml LB, [20ul chlor and 2.5 ul amp] or [20ul tet and 2.5 ul amp]
- Results
- growth in all experimental tubes and no growth in control tubes
10-07-2010
Minipreps
4 tubes of tet in pSB1A3 were prepared and 7 tubes of chlor in pSB1A3 were prepared.
12-07-2010
Restriction Digest with EcoRI Protocol to determine plasmid size:
- pSB1A3 with chlor or tet insert 1ul
- Fast Digest EcoRI 1ul
- 10X Fast Digest Buffer 2ul
- MilliQ H2O 16ul
- Total 20ul
- Incubated at 37C for 5 mins.
Gel Electrophoresis
14-07-2010
Double Digest of chlor, kan and tet in PSB1A3 and amp is pSB1C3 with Xba and PstI protocol
- plasmid with antibiotic part 1ul
- 10X NEBuffer 3 1ul
- MilliQ H20 5ul
- XbaI 1ul
- PstI 1ul
- 10X BSA 1ul
- Total 10ul
- Incubated at 37C for 1 hour.
Gel Electrophoresis
15-07-2010
- Antibiotic parts are now stored in the DNA box according to the BioBytes registry.
- Glycerol stocks were made of overnights from chlor, tet, and kan in PSB1A3 plates.
| Notebook | Building Parts | Testing Parts | Assembly Method | Competent Cells | Plates | Kit Manual | Software |
|---|
 "
"