Team:Aberdeen Scotland/Parts
From 2010.igem.org
(→Part:BBa_K385002: Phage MS2 coat protein) |
I.stansfield (Talk | contribs) |
||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Aberdeen_Scotland/css}} | {{:Team:Aberdeen_Scotland/css}} | ||
{{:Team:Aberdeen_Scotland/Title}} | {{:Team:Aberdeen_Scotland/Title}} | ||
| - | + | <html> | |
| + | <h1>Parts Submitted to the Registry</h1> | ||
| + | </html> | ||
== '''[http://partsregistry.org/Part:BBa_K385002 Part:BBa_K385002]: Phage MS2 coat protein''' == | == '''[http://partsregistry.org/Part:BBa_K385002 Part:BBa_K385002]: Phage MS2 coat protein''' == | ||
| Line 8: | Line 10: | ||
'''Part type''': coding | '''Part type''': coding | ||
| + | |||
'''Part information''' | '''Part information''' | ||
| + | |||
This sequence encodes the MS2 coat protein from phage MS2. It has the property of being able to bind RNA stem loops in a sequence-specific manner. The sequence of the MS2 stem loops is provided in part number BBa_K385000. The coding sequence is supplied without a stop codon, so that it can be used as part of an N-terminal fusion. [http://partsregistry.org/cgi/partsdb/dna.cgi?part_name=BBa_K385002 Sequence analysis] has been confirmed. | This sequence encodes the MS2 coat protein from phage MS2. It has the property of being able to bind RNA stem loops in a sequence-specific manner. The sequence of the MS2 stem loops is provided in part number BBa_K385000. The coding sequence is supplied without a stop codon, so that it can be used as part of an N-terminal fusion. [http://partsregistry.org/cgi/partsdb/dna.cgi?part_name=BBa_K385002 Sequence analysis] has been confirmed. | ||
| - | '''Sequence | + | |
| + | '''Sequence''' | ||
| + | |||
| + | Atggcttctaactttactcagttcgttctcgtcgacaatggcggaactggcgacgtgactgtcgccccaagcaacttcgctaacggggtcgctgaatggatcagctctaactcgcgttcacaggcttacaaagtaacctg | ||
tagcgttcgtcagagctctgcgcagaatcgcaaatacaccatcaaagtcgaggtgcctaaagtggcaacccagactgttggtggagtagagcttcctgtagccgcatggcgttcgtacttaaatatggaactaaccattc | tagcgttcgtcagagctctgcgcagaatcgcaaatacaccatcaaagtcgaggtgcctaaagtggcaacccagactgttggtggagtagagcttcctgtagccgcatggcgttcgtacttaaatatggaactaaccattc | ||
caattttcgctactaattccgactgcgagcttattgttaaggcaatgcaaggtctcctaaaagatggaaacccgattccctcagcaatcgcagcaaactccggcatctacggtgacggtgctggtttaattaac | caattttcgctactaattccgactgcgagcttattgttaaggcaatgcaaggtctcctaaaagatggaaacccgattccctcagcaatcgcagcaaactccggcatctacggtgacggtgctggtttaattaac | ||
| + | |||
'''Design Notes''' | '''Design Notes''' | ||
| + | |||
We omitted the stop codon so this part could be used in a protein fusion construct, with the MS2 protein forming the N-terminal domain. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC) | We omitted the stop codon so this part could be used in a protein fusion construct, with the MS2 protein forming the N-terminal domain. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC) | ||
| + | |||
'''Source ''' | '''Source ''' | ||
| Line 27: | Line 37: | ||
'''Part type''': coding | '''Part type''': coding | ||
| + | |||
| + | |||
| + | '''Part information''' | ||
N-peptide from phage lambda. This protein coding sequence functions in a phage transcriptional termination control mechanism, by binding to an RNA stem loop (B-box [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385005 Part:BBa_K385005]) in a sequence specific manner. This peptide can be used as part of a translational control strategy for eukaryote gene expression. The B-box sequence should be placed in the 5' leader of a gene whose expression is to be controlled, and the N-peptide is expressed in trans to regulate ribosomal scanning. [http://partsregistry.org/cgi/partsdb/dna.cgi?part_name=BBa_K385003 Sequence analysis] has been confirmed. | N-peptide from phage lambda. This protein coding sequence functions in a phage transcriptional termination control mechanism, by binding to an RNA stem loop (B-box [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385005 Part:BBa_K385005]) in a sequence specific manner. This peptide can be used as part of a translational control strategy for eukaryote gene expression. The B-box sequence should be placed in the 5' leader of a gene whose expression is to be controlled, and the N-peptide is expressed in trans to regulate ribosomal scanning. [http://partsregistry.org/cgi/partsdb/dna.cgi?part_name=BBa_K385003 Sequence analysis] has been confirmed. | ||
| - | '''Sequence | + | |
| + | '''Sequence''' | ||
atggatgctcaaactagaagaagagaaagaagagctgaaaaacaagctcaatggaaagctgctaatggtgacggtgctggtttaattaac | atggatgctcaaactagaagaagagaaagaagagctgaaaaacaagctcaatggaaagctgctaatggtgacggtgctggtttaattaac | ||
| + | |||
'''Applications''' | '''Applications''' | ||
| - | The Aberdeen 2010 iGEM team has no direct experience of using [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385003 BBa_K385003], but the closely related part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385004 BBa_K385004]. consisting of a tandem repeat of the N-peptide, allowed the functional expression of a downstream GFP reporter. | + | The Aberdeen 2010 iGEM team has no direct experience of using [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385003 BBa_K385003], but the closely related part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385004 BBa_K385004]. consisting of a tandem repeat of the N-peptide, allowed the functional expression of a downstream GFP reporter. |
| + | |||
'''Design Notes''' | '''Design Notes''' | ||
The part was engineered with an AUG, but no stop codon, to allow the part to be used as a translational fusion with another downstream open reading frame. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC) | The part was engineered with an AUG, but no stop codon, to allow the part to be used as a translational fusion with another downstream open reading frame. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC) | ||
| + | |||
| + | |||
| + | '''Source ''' | ||
| + | Phage lambda genome | ||
| + | |||
| + | == '''[http://partsregistry.org/Part:BBa_K385004 Part:BBa_K385004]: Phage lambda N-peptide, tandem repeat == | ||
| + | |||
| + | '''Length''': 177 bp | ||
| + | |||
| + | '''Part type''': coding | ||
| + | |||
| + | |||
| + | '''Part information''' | ||
| + | |||
| + | Two copies of the N-peptide from phage lambda, arranged as a tandem repeat. The N-peptide protein coding sequence functions in a phage transcriptional termination control mechanism, by binding to an RNA stem loop (B-box) in a sequence specific manner. This peptide can be used as part of a translational control strategy for eukaryote gene expression. The B-box sequence should be placed in the 5' leader of a gene whose expression is to be controlled, and the N-peptide is expressed in trans to regulate ribosomal scanning. Tandem repeats of the N-peptide were cloned in this BioBrick so as to optimise binding opportunities to the target mRNA stem loop. [http://partsregistry.org/cgi/partsdb/dna.cgi?part_name=BBa%20K385004 confirmed sequence] | ||
| + | |||
| + | |||
| + | '''Sequence''' | ||
| + | |||
| + | atggatgctcaaactagaagaagagaaagaagagctgaaaaacaagctcaatggaaagctgctaatggtgacggtgctggtttaattaacgacgctcaaa | ||
| + | cccgtagaagagagagaagagccgaaaagcaagctcaatggaaggccgctaacggtgatggcgccggcttgattaat | ||
| + | |||
| + | |||
| + | '''Applications''' | ||
| + | |||
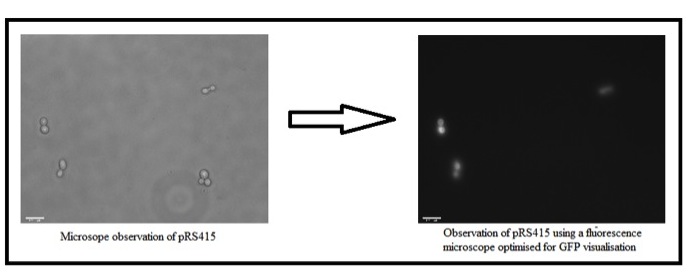
| + | The N-peptide tandem repeat reading frame was fused in-frame to GFP to make a translational fusion. It was placed under control of the yeast GAL1 promoter (BBa_J63006), and transformed into yeast Saccharomyces cerevisiae in the single copy shuttle vector pRS415. | ||
| + | The transformants were grown overnight in synthetic defined medium containing 2% w/v galactose, and observed using a fluorescence microscope optimised for GFP visualisation (Figure 1). | ||
| + | |||
| + | [[Image:PRS415_FLU.jpg|center|800 px]] | ||
| + | |||
| + | A control culture of the same transformant was grown using glucose as the carbon source; these conditions do not activate the GAL promoter. The results (Figure 2) show no GFP fluorescence. | ||
| + | Overall the results indicate that the N-peptide can be successfully expressed as a protein fusion with other standard parts. | ||
| + | |||
| + | |||
| + | '''Design Notes''' | ||
| + | |||
| + | The part was engineered with an AUG, but no stop codon, to allow the part to be used as a translational fusion with another downstream open reading frame. A glycine rich spacer peptide was inserted at the 3' end of each of the tandem N-peptide repeats, to allow the N-peptide to be separated from each other, and any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC) | ||
| + | |||
| + | '''Source ''' | ||
| + | Phage lambda genome | ||
| + | |||
| + | == '''[http://partsregistry.org/Part:BBa_K385005 Part:BBa_K385005]: B-box sequence encoding a regulatory mRNA stem loop == | ||
| + | |||
| + | '''Length''': 56 bp | ||
| + | |||
| + | '''Part type''': Regulatory | ||
| + | |||
| + | |||
| + | '''Part information''' | ||
| + | |||
| + | This part encodes a sequence that is capable of forming a stem loop in the mRNA. Moreover, this stem loop is bound in a sequence and structure-specific manner by the N-peptide sequence (see part numbers [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385003 BBa_K385003] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K385004 BBa_K385004]). The mRNA stem sequence is derived from phage lambda, and forms part of a transcriptional termination attenuation system. This stem loop encoding sequence can be used as part of a eukaryote gene expression control strategy. Insertion of this stem loop into the 5' untranslated region (5'UTR) of a target gene (i.e. between the transcript start site and the AUG translation initiation site) will allow this mRNA to be actively translated in the absence of the N-peptide sequence. However, expression of the N-peptide in trans will allow N-peptide binding to the B-box stem, causing translational attenuation by inhibition of ribosome scanning along the 5'UTR. | ||
| + | |||
| + | |||
| + | '''Sequence''' | ||
| + | |||
| + | attatctacttaagggccctgaagaagggcccttaagaacacaaaattcgagacat | ||
| + | |||
'''Source ''' | '''Source ''' | ||
Phage lambda genome | Phage lambda genome | ||
| + | <html> | ||
| + | <br><br> | ||
| + | <hr> | ||
| + | <table class="nav"> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <a href="https://2010.igem.org/Team:Aberdeen_Scotland/Protocols"><img src="https://static.igem.org/mediawiki/2010/8/8e/Left_arrow.png"> Return to Protocols</a> | ||
| + | </td> | ||
| + | <td align="right"> | ||
| + | <a href="https://2010.igem.org/Team:Aberdeen_Scotland/Modeling">Continue to the Modelling Summary <img src="https://static.igem.org/mediawiki/2010/3/36/Right_arrow.png"></a> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | {{:Team:Aberdeen_Scotland/Footer}} | ||
Latest revision as of 15:57, 27 October 2010
University of Aberdeen - ayeSwitch
Parts Submitted to the Registry
Contents |
Part:BBa_K385002: Phage MS2 coat protein
Length: 414 bp
Part type: coding
Part information
This sequence encodes the MS2 coat protein from phage MS2. It has the property of being able to bind RNA stem loops in a sequence-specific manner. The sequence of the MS2 stem loops is provided in part number BBa_K385000. The coding sequence is supplied without a stop codon, so that it can be used as part of an N-terminal fusion. Sequence analysis has been confirmed.
Sequence
Atggcttctaactttactcagttcgttctcgtcgacaatggcggaactggcgacgtgactgtcgccccaagcaacttcgctaacggggtcgctgaatggatcagctctaactcgcgttcacaggcttacaaagtaacctg tagcgttcgtcagagctctgcgcagaatcgcaaatacaccatcaaagtcgaggtgcctaaagtggcaacccagactgttggtggagtagagcttcctgtagccgcatggcgttcgtacttaaatatggaactaaccattc caattttcgctactaattccgactgcgagcttattgttaaggcaatgcaaggtctcctaaaagatggaaacccgattccctcagcaatcgcagcaaactccggcatctacggtgacggtgctggtttaattaac
Design Notes
We omitted the stop codon so this part could be used in a protein fusion construct, with the MS2 protein forming the N-terminal domain. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC)
Source
see NCBI sequence
Part:BBa_K385003: Phage lambda N-peptide
Length: 90 bp
Part type: coding
Part information
N-peptide from phage lambda. This protein coding sequence functions in a phage transcriptional termination control mechanism, by binding to an RNA stem loop (B-box Part:BBa_K385005) in a sequence specific manner. This peptide can be used as part of a translational control strategy for eukaryote gene expression. The B-box sequence should be placed in the 5' leader of a gene whose expression is to be controlled, and the N-peptide is expressed in trans to regulate ribosomal scanning. Sequence analysis has been confirmed.
Sequence
atggatgctcaaactagaagaagagaaagaagagctgaaaaacaagctcaatggaaagctgctaatggtgacggtgctggtttaattaac
Applications
The Aberdeen 2010 iGEM team has no direct experience of using BBa_K385003, but the closely related part BBa_K385004. consisting of a tandem repeat of the N-peptide, allowed the functional expression of a downstream GFP reporter.
Design Notes
The part was engineered with an AUG, but no stop codon, to allow the part to be used as a translational fusion with another downstream open reading frame. A glycine rich spacer peptide was inserted at the 3' end of the sequence, to allow the N-peptide to be separated from any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC)
Source
Phage lambda genome
Part:BBa_K385004: Phage lambda N-peptide, tandem repeat
Length: 177 bp
Part type: coding
Part information
Two copies of the N-peptide from phage lambda, arranged as a tandem repeat. The N-peptide protein coding sequence functions in a phage transcriptional termination control mechanism, by binding to an RNA stem loop (B-box) in a sequence specific manner. This peptide can be used as part of a translational control strategy for eukaryote gene expression. The B-box sequence should be placed in the 5' leader of a gene whose expression is to be controlled, and the N-peptide is expressed in trans to regulate ribosomal scanning. Tandem repeats of the N-peptide were cloned in this BioBrick so as to optimise binding opportunities to the target mRNA stem loop. confirmed sequence
Sequence
atggatgctcaaactagaagaagagaaagaagagctgaaaaacaagctcaatggaaagctgctaatggtgacggtgctggtttaattaacgacgctcaaa cccgtagaagagagagaagagccgaaaagcaagctcaatggaaggccgctaacggtgatggcgccggcttgattaat
Applications
The N-peptide tandem repeat reading frame was fused in-frame to GFP to make a translational fusion. It was placed under control of the yeast GAL1 promoter (BBa_J63006), and transformed into yeast Saccharomyces cerevisiae in the single copy shuttle vector pRS415. The transformants were grown overnight in synthetic defined medium containing 2% w/v galactose, and observed using a fluorescence microscope optimised for GFP visualisation (Figure 1).
A control culture of the same transformant was grown using glucose as the carbon source; these conditions do not activate the GAL promoter. The results (Figure 2) show no GFP fluorescence. Overall the results indicate that the N-peptide can be successfully expressed as a protein fusion with other standard parts.
Design Notes
The part was engineered with an AUG, but no stop codon, to allow the part to be used as a translational fusion with another downstream open reading frame. A glycine rich spacer peptide was inserted at the 3' end of each of the tandem N-peptide repeats, to allow the N-peptide to be separated from each other, and any downstream ORF by a flexible linker. (Linker sequence GGT GAC GGT GCT GGT TTA ATT AAC)
Source Phage lambda genome
Part:BBa_K385005: B-box sequence encoding a regulatory mRNA stem loop
Length: 56 bp
Part type: Regulatory
Part information
This part encodes a sequence that is capable of forming a stem loop in the mRNA. Moreover, this stem loop is bound in a sequence and structure-specific manner by the N-peptide sequence (see part numbers BBa_K385003 and BBa_K385004). The mRNA stem sequence is derived from phage lambda, and forms part of a transcriptional termination attenuation system. This stem loop encoding sequence can be used as part of a eukaryote gene expression control strategy. Insertion of this stem loop into the 5' untranslated region (5'UTR) of a target gene (i.e. between the transcript start site and the AUG translation initiation site) will allow this mRNA to be actively translated in the absence of the N-peptide sequence. However, expression of the N-peptide in trans will allow N-peptide binding to the B-box stem, causing translational attenuation by inhibition of ribosome scanning along the 5'UTR.
Sequence
attatctacttaagggccctgaagaagggcccttaagaacacaaaattcgagacat
Source
Phage lambda genome
 Return to Protocols Return to Protocols
|
Continue to the Modelling Summary 
|
 "
"