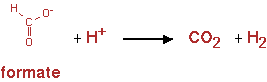Team:TU Delft/Modeling/MFA/additional pathways
From 2010.igem.org
Pathways added to the E. coli metabolic network
The E. coli network from CellNetAnalyzer contains, glycolysis, TCA cycle, pentose phosphate pathway, gluconeogenesis, anapleorotic routes, oxydative phosphorilization and biosynthesis pathways. CellNetAnalyzer can be found [http://www.mpi-magdeburg.mpg.de/projects/cna/cna.html here] with this network.
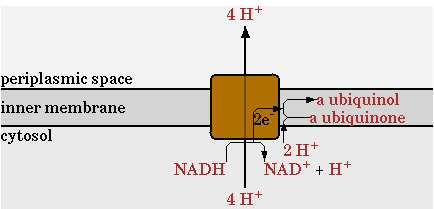
There was one change made in the network provided by CellNetAnalyzer for the NADH dehydrogenase reaction, annotated as NADHdehydro in CellNetAnalyzer. In the network of CellNetAnalyzer this reaction exports two protons, but this was changed to 4 as given by Ecocyc. This reaction is shown in figure 1.
Alkane degradation
To link alkanes to the existing network, the beta-oxidation was chosen as entry point. The reactions from several genes from biobricks were used to transform alkanes in to fatty acids which enter the beta oxidation cycle. These genes are listed below. The reactions used for CNA already leave out some redundant information. For example only NADH is used instead of NADH and NAD+.
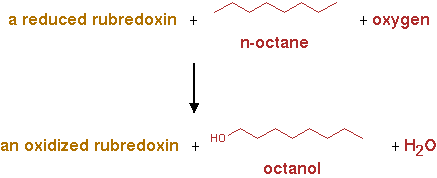
AlkB2 (EC 1.14.15.3)
The reaction for AlkB2 is shown in figure 2.
The reaction was implemented in CNA as;
n-alkane + reduced rubredoxin + O2 -> n-alkanol + oxidized rubredoxin
RubA3/RubA4 (EC 1.18.1.1)
The reaction for the regeneration of rubredoxin is shown in figure 3.
The reaction was implemented in CNA as;
oxidized rubredoxin + NADH -> reduced rubredoxin
This and the previous reaction are performed by this [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398014 BioBrick] and this [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398017 BioBrick]
ADH (EC 1.1.1.1)
The reaction for ADH is shown in figure 4.
The reaction was implemented in CNA as;
n-alkanol -> n-aldehyde + NADH
This reaction is performed by this [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398018 BioBrick]
ALDH (EC 1.2.1.3)
The reaction for ALDH is shown in figure 5.
The reaction was implemented in CNA as;
n-aldehyde -> n-fatty acid + NADH
This reaction is performed by this [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398029 BioBrick]
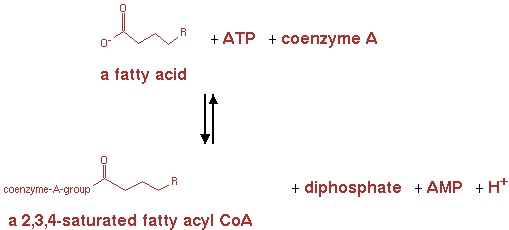
fatty acyl-CoA synthetase (EC 6.2.1.3)
From here the genes are already present in the E. coli genome. They were not yet present in the metabolic network for E. coli in CellNetAnalyzer however. So the following reactions and pathways were also implemented in CNA. The reaction for fatty acyl-CoA synthetase is shown in figure 6.
The reaction was implemented in CNA as;
n-fatty acid + ATP -> n-saturated fatty acyl-CoA + AMP
Adenylate kinase (EC 2.7.4.3)
CNA did not have a reaction to regenerate AMP yet, so the reaction for adenylate kinase was added to the network. The reaction for adenylate kinase is shown in figure 7.
The reaction was implemented in CNA as;
ATP + AMP -> 2 ADP
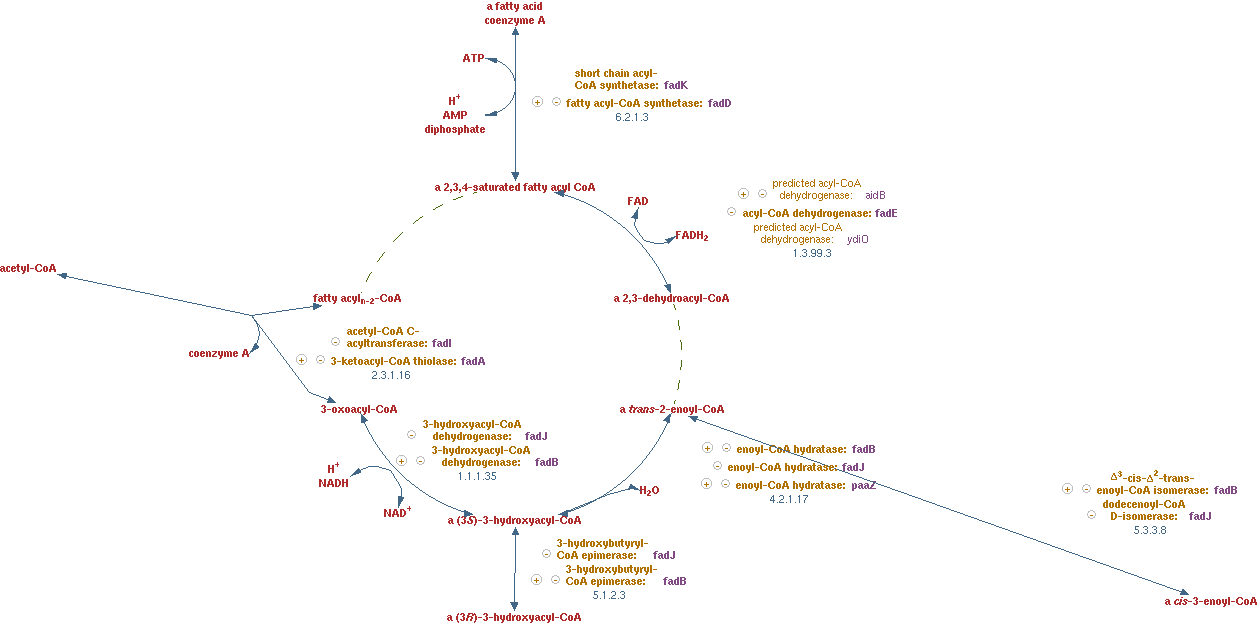
Fatty acid beta-oxidation cycle (EC 1.3.99.3 EC 4.2.1.17 EC 1.1.1.35 EC 2.3.1.16)
The reactions for this pathway are shown in figure 8.
The lumped reaction was implemented in CNA as;
n-saturated fatty acyl-CoA -> (n - 2)-saturated fatty acyl-CoA + acetyl-CoA + FADH2 + NADH
FADH2 regeneration
The cofactor FADH2 is not yet defined in the network of CNA, so a reaction had to be introduced to regenerate it. FADH2 gives its electrons to ubiquinol just like NADH, however in this process no protons are exported.
The reaction was implemented in CNA as;
1 FADH2 -> 1 QH2
Odd numbered alkanes
For all even numbered alkanes the above reactions completely link them to the main network in CNA. All the co-factors have regeneration reactions and all the alkanes are converted into acetyl-CoA. The final reaction in the beta oxidation cycle (n = 4) produces then two acetyl-CoA. For odd numbered alkanes the final reaction however (n = 5), a propionyl-CoA is generated;
n-saturated fatty acyl-CoA -> propionyl-CoA + acetyl-CoA + FADH2 + NADH
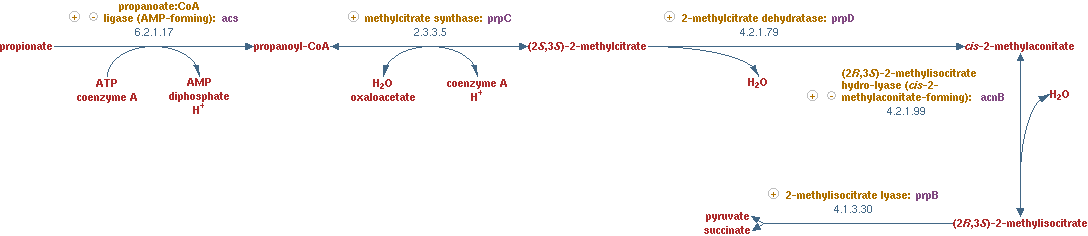
2-methylcitrate cycle (EC 2.3.3.5 EC 4.2.1.79 EC 4.2.1.99 EC 4.1.3.30)
This propionyl-CoA still needs to be linked to the main network in CNA. The pathway that was used to process this metabolite, was a part of the 2-methylcitrate cycle. The reactions for this pathway are shown in figure 9.
The lumped reaction was implemented in CNA as;
propionyl-CoA + oxaloacetic acid -> succinate + pyruvate
Biomass formation
The biomass is formed by many anabolic reactions that make monomers. All the anabolic reactions start at the so called key metabolites. There are 12 key metabolites and they are all in the glycolytic pathway and the TCA cycle. In the tool they are the red metabolites. For this analysis the anabolic reactions already present in CNA were used. These are a lot of reactions however and they will not be shown in this analysis, although they do occur. In the tool growth can ben seen as consumption of the key metabolites.
NO3 as electron acceptor
In oily environments oxygen diffuses more difficult into the water phase. The oxygen is used for the oxidative phosphorylation, regenerating NADH, and for the first step in the hydrocarbon degradation. To be more efficient with oxygen an additional electron acceptor was introduced.
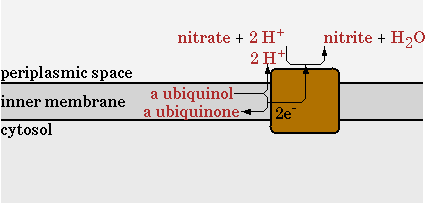
The standard oxidative phosphorylation (EC 1.6.5.3 EC 1.10.2.-) is shown in figure 10.
The second step will be disabled in the network and be replaced with a nitrate reductase (EC 1.7.99.4). This reaction is shown in figure 11.
The reaction was implemented in CNA as;
NO3- + QH2 -> NO2-
This reaction uses NO3 as an electron acceptor to regenerate NADH and export protons to generate ATP. Less protons are exported per mol of NADH in comparison with oxygen as electron acceptor, so the ATP/NADH ratio will drop. The goal of implementing this pathway however, is to see how much the oxygen requirement of E. coli can maximally be reduced.
PHB production
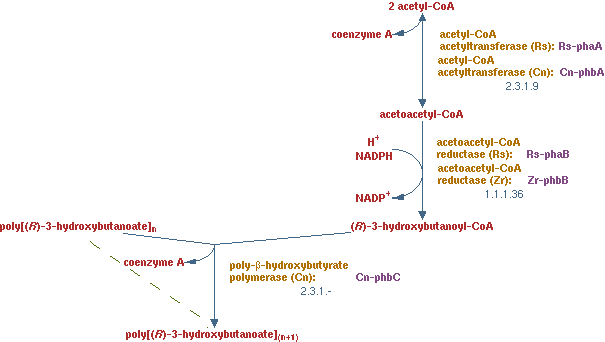
In previous situations the hydrocarbons were degraded only to form biomass and CO2. It is interesting to see how much product theoretically could be made from hydrocarbons. PHB is a polymer of polyhydroxybutyrate. The production pathway of PHB is well known. PHB is a solid product which is easy to recover in the down stream process.
The pathway is displayed in figure 12. The enzymes present in this pathway are EC 2.3.19, EC 1.1.1.36 and EC 2.3.1.-.
The lumped reaction was implemented in CNA as;
2 acetyl-CoA + NADPH -> (R)-3-hydroxybutanoyl-CoA
the polymerization reaction just consumes (R)-3-hydroxybutanoyl-CoA, because a solid has no concentration in the liquid and therefore does not need to fulfill the steady-state condition. There are limiting factors to this reaction, but those are not considered in this analysis.
Hydrogen production
As with PHB, this pathway was added to the metabolic pathway to see how much product could be made from the alkane degradation in stead of just biomass and CO2 formation. Hydrogen is considered a green fuel as its combustion produces water. Hydrogen is a volatile product and is also easily separated from fermentation broth. It does however contain no carbon atoms, so hydrogen production will still result in production of CO2 and biomass.
The pathway is shown in figure 13
The reaction was implemented in CNA as;
Formate -> H2 + CO2
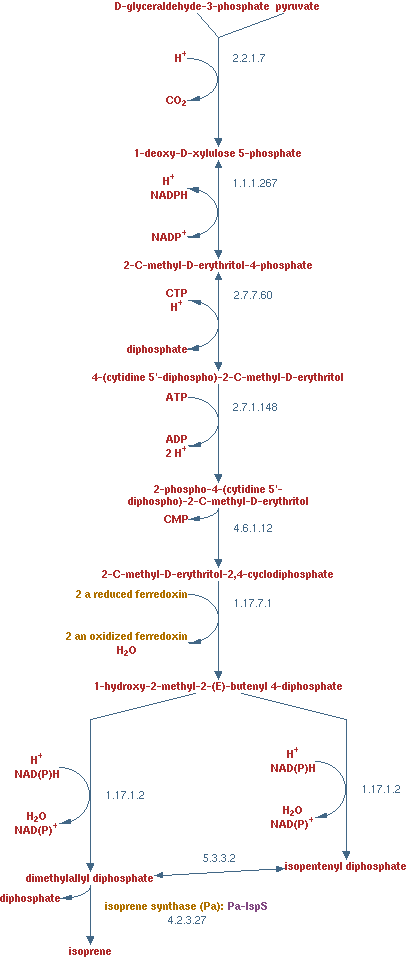
Isoprene production
As with the previous product pathways, isoprene is a product that can easily be separated from liquid. Isoprene is a volatile substance found in plants. E. coli will not be able to produce is in the near future, but it is an interesting product. It is a very reduced product, with a similar amount of electron per carbon atom as alkanes. Alkanes have 6 - 6.3 electrons per carbon atom depending on the length and isoprene has 5.6 electron per carbon electron.
The pathway is displayed in figure 14. The enzymes in the pathway are EC 2.2.1.7, EC 1.1.1.267, EC 2.7.7.60, EC 2.7.1.148, EC 4.6.1.12, EC 1.17.7.1, EC 1.17.1.2 and EC 4.2.3.27. The reaction of enzyme EC 1.17.7.1 has been adjusted to generate only 1 oxidized ferredoxin. This was done to hold the electron balance. Ferredoxin is regenerated by consuming a NADH.
The lumped reaction was implemented in CNA as;
1 pyruvate + 1 D-glyceraldehyde-3-phosphate + 1 NADPH + 2 NADH + 3 ATP -> isoprene + CO2
Go to the detailed results of the metabolic flux analysis here
Go back the metabolic flux analysis main page here
.
 "
"