Team:HokkaidoU Japan/Notebook/August30
From 2010.igem.org
Digestion of pUC119 by EcoR I, Pst I
| - | EcoR I | Pst I | E, P | Old EcoR I | |
| DNA solution | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| DW | 17 uL | 14 uL | 14 uL | 13 uL | 14 uL |
| 10x M buffer | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL |
| 0.1% BSA | - | 2 uL | 2 uL | 2 uL | 2 uL |
| EcoR I | - | 1 uL | - | 1 uL | 1 uL |
| Pst I | - | - | 1 uL | 1 uL | - |
| Total | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL |
→Incubated at 37C for 60 min →Electrophoresed 2 uL for confirmation
- There were no bands, forgot to add DNA
- Reused the remaining 18 uL of digestion solution by adding 1 uL of ADW and 1uL of DNA
→Incubated at 37C for 60 min
→Electrophoresed 2 uL of each solution(+ 0.4 uL 6x SB)
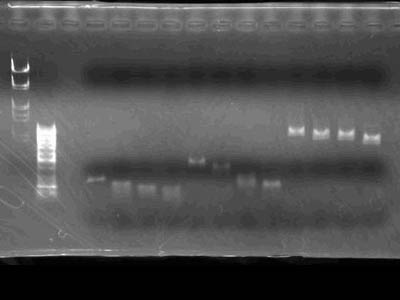
Electrophoresis
| Lane | DNA |
| 2 | λ/HindIII, EcoR I(4 uL used) |
| 3 | Undigested |
| 4 | EcoR I |
| 5 | Pst I |
| 6 | EcoR I + Pst I |
| 7 | EcoR I (used old enzyme to check it's activity) |
- In lane 3 monomers, dimers and trimers of plasmid were visible .
- From lanes 4 through 7 it's visible that DNA digestion wasn't satisfactory
- Because plasmid became linear it's was slower than super-coiled one's
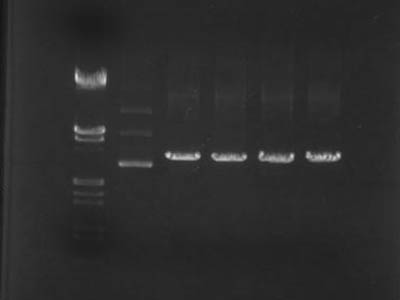
Digestion of parts PCRed using digestion visualization primers
| RBS | double terminator | GFP | ||||||||||
| DNA | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| DW | 17 uL | 14 uL | 14 uL | 13 uL | 17 uL | 14 uL | 14 uL | 13 uL | 17 uL | 14 uL | 14 uL | 13 uL |
| 10x M buffer | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL |
| 0.1% BSA | - | 2 uL | 2 uL | 2 uL | - | 2 uL | 2 uL | 2 uL | - | 2 uL | 2 uL | 2 uL |
| EcoR I | - | 1 uL | - | 1 uL | - | 1 uL | - | 1 uL | - | 1 uL | - | 1 uL |
| Pst I | - | - | 1 uL | 1 uL | - | - | 1 uL | 1 uL | - | - | 1 uL | 1 uL |
| Total | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL | 20 uL |
→Incubated at 37C for 60 min
Electrophoresis
- Markers used λ/HindIII, EcoR I and 50bp ladder
- Was obvious that parts were cut as intended
 "
"