Team:ETHZ Basel/Biology/Implementation
From 2010.igem.org
Implementation
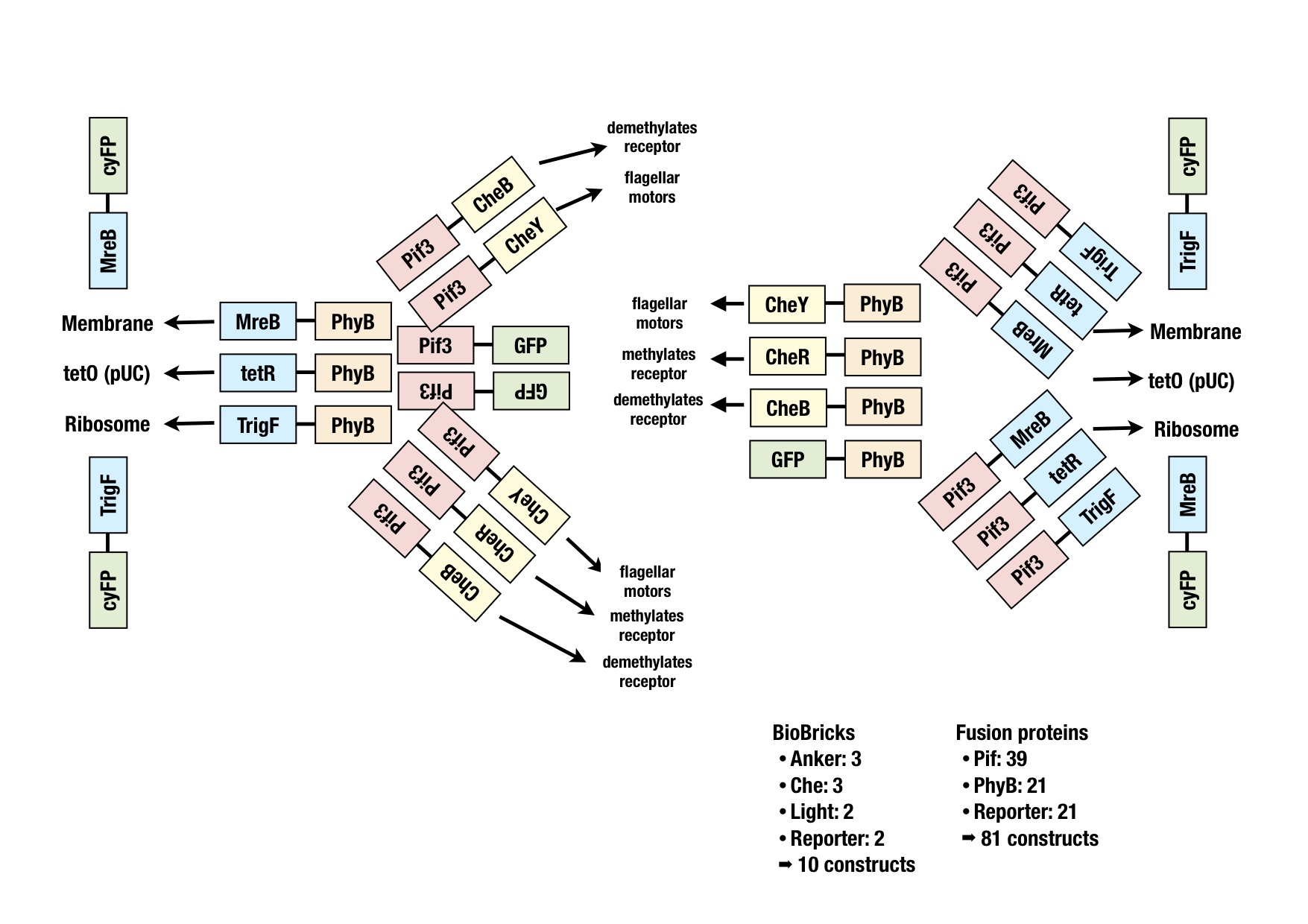
generation of fusion proteins
The image shows all the fusion proteins that were planned. The BioBricks for all these constructs were generated via PCR or synthesized at GeneArt and ligated into the storage vector for sequencing. Easy cutting and pasting into working vectors should be possible with the cloning strategy BBF RFC28 (http://dspace.mit.edu/handle/1721.1/46721).
Modeller's input
Due to the high amount of fusion proteins that were in planing (81 constructs), priorities had to be distributed to the different genes. This was possible due to the various models the dry-lab team implemented.
- Chemotactic protein: CheY was chosen as first target.
- Anchor: TetR was the first choice due to its wide application in synthetic biology and extensive characterization.
- Pif3 linked to Che-Protein: Although PhyB seemed to be the more robust choice, Pif3 was chosen to be linked to the Che-Protein due to the tendency of PhyB to sequester.
- Ratio anchor to binding partner: The simulations favoured a ratio of 50 µM anchor to 40 µM anchor binding partner.
Plasmid copy number
As the ratio between the amount of anchor and its binding partner has been shown to be essential (Team:ETHZ_Basel/Modeling/Collaboration), the plasmid copy number was determined by normalization of cell number via optical density measurement followed by plasmid concentration measurements using a commercial Miniprep kit. The results clearly show that a concentration of tetO = 50 µM in one cell can not be achieved even by a high copy number plasmid such as pUC19. Therefore, the decision of integrating a second anchor protein also fused to the light binding protein PhyB was made.
Functionality assays
The constructs will have to be tested for the following properties:
- Che protein fusion: Using the chemotactic assay described by Mazumder et al., the functionality of Che protein fusions can be tested.
- Localizer fusion: Spatial localization of the anchor protein to either the plasmid (tetR-tetO), the cell membrane (mreB) or the ribosome (trigA) can be investigated by fusing the anchor binding protein to a fluorescent protein.
- PhyB-Pif3 system: Fusing a second fluorescent protein to Pif3 would enable the visualization of the light activated dimerization.
 "
"