Team:RMIT Australia/Notebook
From 2010.igem.org


Notebook
|
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
20 AugustDid two PCR reaction and a transformation The first PCR we ran was a touch down PCR. Temperatures ranged from 69 to 64. The second PCR was a two step PCR. 30 AugustOrdered the TAQ polymerase from Mr. Gene. Should have it in 15 days. Meeting today at 10am to discuss project budget and costings. After the meeting we all are sitting together for a writing session. 31 AugustSpent the morning organising our flights In the afternoon we all sat together for a writing session 1 SeptemberWe all sat together for another writing session. 7 SeptemberHad a team meeting 14 SeptemberHad a team meeting and made a lab time table, we also set out tasks which need to be done by the next meeting
15 SeptemberMutagenesis Quikchange XL kit Click Here for Mutagenesis Protocol
We optimised for low 260/280 Reaction Mixture
First mutagenesis was to turn the -35 site in the promoter region into an Afi II restriction site. Dpn1 digest (2 µl) for 2hrs 16 SeptemberClick Here for Mutagenesis Protocol 2 µl of the mutagenesis product into 100 µl of competent cells Plate 50 µl, 100 µl and 150 µl samples. 17 September- finalized project abstract 19 September- Sent off project abstract to HQ 20 SeptemberTwo colonies were picked from the plates and were grown in 5mL of Media 21 SeptemberA miniprep was performed on 1.5ul of the overnight culture. 10ng/ul DNA were obtained from both samples with a O.D higher than 1.85. These samples were then used to perform a restriction digest using EcoR1 H.F and AflII. The restriction digest was done according to the iGEM protocols [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.pdf CLICK HERE]. Results The restriction digest did not work. We expected a band at 160bp and one at ~3000bp 22 September>>>> TABLE <<<<
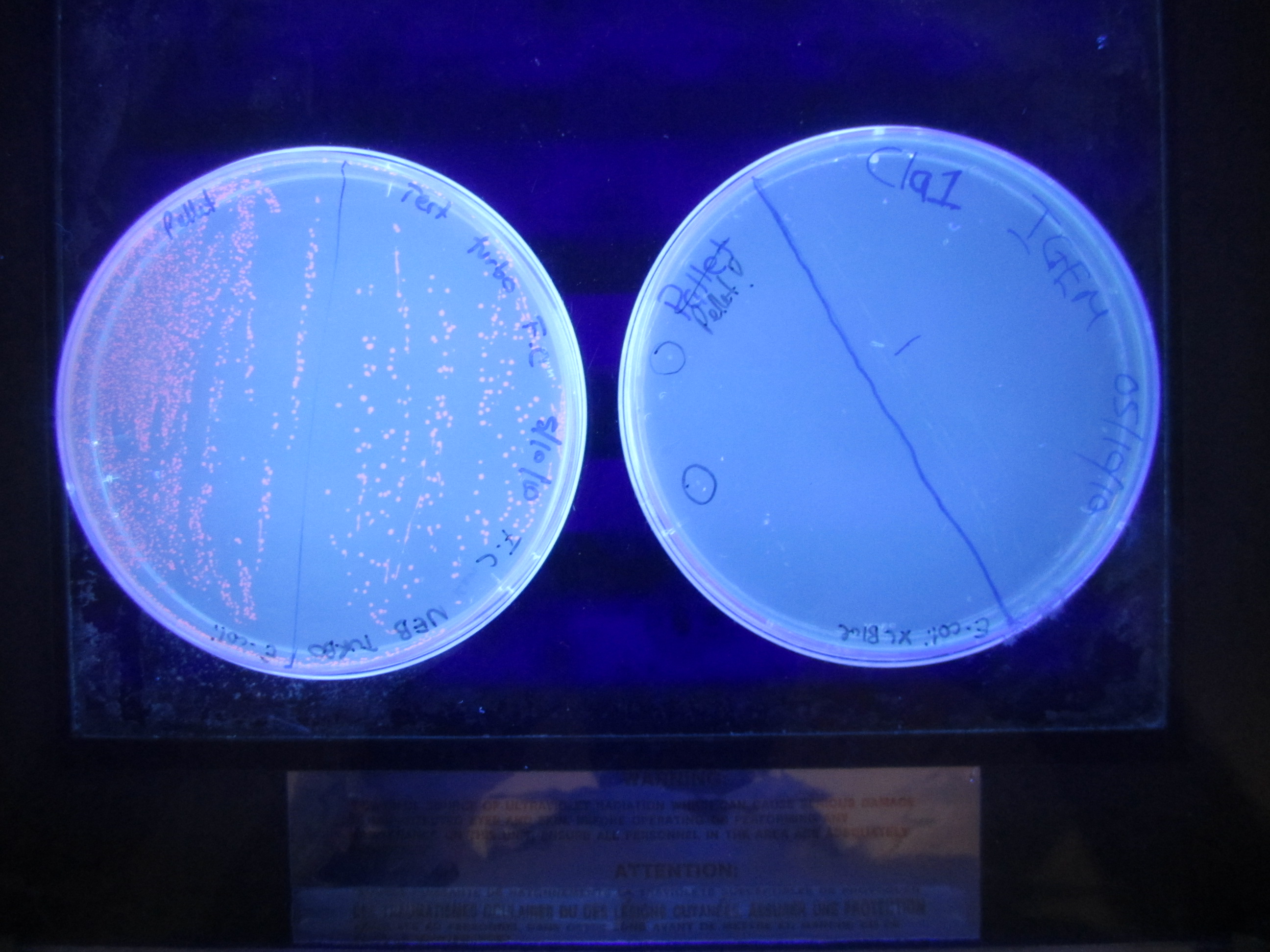
23 SeptemberSecond mutagenesis. This involves changing the -10 promoter region into a ClaI restriction site. AflII (11) miniprep sample was used as template Click Here for Mutagenesis Protocol After the Dpn1 digest, 2ul of product was transformed into E.coli Xl1 blue cells. 24 SeptemberColonies grew on the plate. This was stores in the fridge (4 degrees) until Monday for colonies to be picked. 27 SeptemberTwo colonies were picked from the plate and grown overnight at 37 degrees in the shaking incubator. 28 SeptemberPreparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. 29 SeptemberTwo more colonies (different) were picked and grown overnight at 37 degrees. Transformation of RFP in Ampicillin, Kanamycin and Chloroamphenicol backbones. They will act as spare backbones. 30 SeptemberA second preparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture again. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. There is a problem with the backbone or the miniprep. Must investigate. 4 OctoberGot it! The problem was the plasmid backbone. It was thought that it was pSB1A3, but in fact it was pSB1AK3. Kanamycin has a ClaI site. A restriction digest was performed using EcoRI and PstI. The RFP plasmid was also digested with EcoRI and PstI. A ligation reaction was performed in order for the promoter part to switch backbones (from pSB1AK3 to pSB1C3). RFP is also a form of control mechanism to see if the plasmid has the correct insert. Red colonies have RFP, while White colonies have our insert. Overnight ligation at 16 degrees. 5 OctoberThe ligation product was transformed (5ul) into E.coli XL1 Blue competent cells. 6 OctoberColonies grew. Two white colonies were picked and grown overnight in the shaking incubator at 37 degrees. |
 "
"