Team:UCL London/Approach
From 2010.igem.org
Biochemical Engineering
As a team, we have a vision that our cells can detect external changes in the environment and hence be programmed to response to them, instead of us manually having to detect and resonse. Our project is based primarily on biochemical engineering processes and it's essential that the audience understands what us Biochemical Engineers are about and what we do, especially as this is unfamiliar territory to many.
In our department, we believe that extraordinary advances in the life sciences have great potential to improve our quality of life, this is ultimately achieved through better medicines and a cleaner environment. We provide the foundations for the transforming of such extraordinary next discoveries into products and processes leading to the improvement of health and welfare state of the nation.
Fermenters
For the first time, UCL iGEM 2010 will be implementing the use of its sophisticated class of 1 litre fermenters. In the biopharmaceutical industry, protein expression takes place on a large scale in these devices although at scales by far larger than the ones to be put into use during this project. The recombinant E.coli cells are grown under a controlled environment within the fermenters whose scales can be as large as 10,000 L. A medium is firstly created containing vital chemicals which will ensure that the cells have the vital requirements to maintain growth Fermentation Protocol.
The diagram shows one of the 1 litre fermenters that will be used to test one of our strains. The main parts have been labeled and are as follows;
Main Fermentation Chamber: This is where our E.Coli cells are grown and where the fermentation process takes place
Impellers: Responsible for ensuring good flow conditions as well as
Temperature probe: Can be used to measure the temperature of the medium.
pH probe: Used to detect any changes in the pH
Base and Acid can be used to adjust the pH o the vessel ensuring ideal conditions
In let air: Air is pumping into the vesel through this and provides the cells with the Oxygen
Compressor
Out let air:
Sampling tube: Samples of the medium can be taken at various intervals and then OD or stored
Inoculation opening: It is through this opening that the initial Inoculum is added
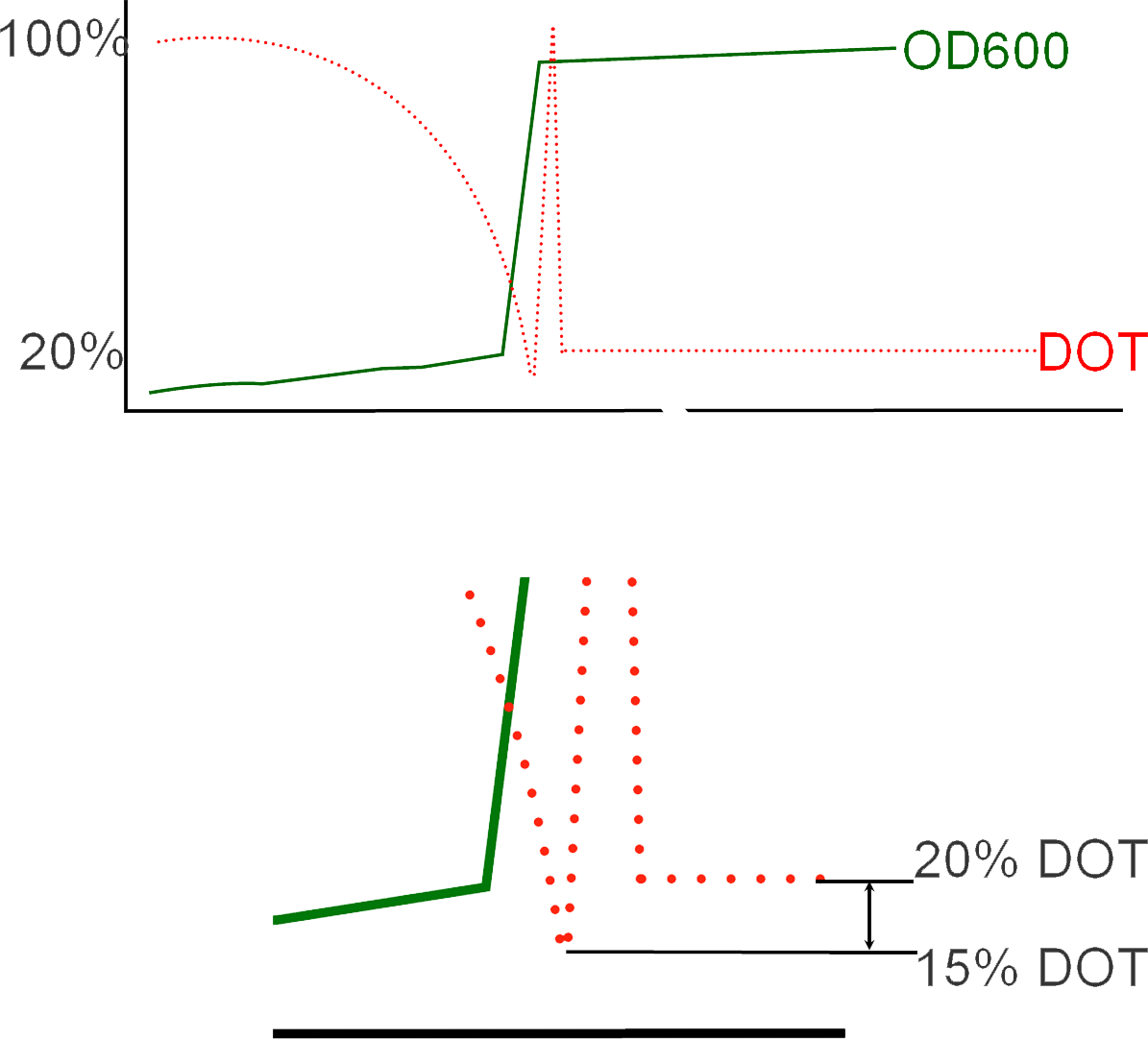
DOT levels during fermentation
During fermentation, initially, DOT levels in the batch fermenter will be observed gradually decreasing as the E.Coli cells multiply and grow. With time, the DOT in the vessel will fall below 20%, at which point the situation becomes critical because maintaining such low levels of oxygen will result in cellular death and loss of batch. And so this is the ideal time to add IPTG and induce protein expression. The addition is followed with the conversion of the process from batch to fed batch as Oxygen is now pumped into the vessel resulting in the shooting up of DOT levels as shown on the graph below. This is also complemented with the addition of glycerol to help maintain the cells during protein expression. The resultant transition from cellular growth to protein expression can be observed in the immediate increase in OD values subsequent to inoculation.
Protein Expression
E.coli is essentially one of the most widely applied in-vivo expression systems. The reason for its wide application is the fact that it is so well developed and the E. coli genome is fully developed and well understood. In biotechnology, as an example, a DNA sequence for a protein of interest could be inserted into a high copy-number plasmid containing the lac promoter, which is then transformed into the bacterium Escherichia coli. Addition of IPTG (a lactose analog) activates the lac promoter and causes the bacteria to express the protein of interest.
One technique of insuring high levels of a protein is to clone the gene downstream of a well-characterized regulated promoter. In our case, we will be using pTAC, the tac promoter, a very widely used expression system. Being the strong hybrid promoter it is, it is repressed by the LacI protein, and on addition of IPTG, the lacI repressser is inactivated. This inactivtation breaks the strong repression of pTAC resulting in expression of pTAC. It has been shown that high expressions of pTAC is directly proportional to the concentration of IPTG added. And so by varying the concentratrion of IPTG added, you can regulate the rate of the expression of the desired protein downstream.
That’s essentially the biology behind protein expression. The engineering side is where our project really gets interesting.
Our Animation
Thanks to the efforts of our artists, we managed to summarize what we hope to achieve in the fermentation unit in a simple animation;
Here, the green blocks represent the level of DOT in the fermenter, red-blue being the rate of protein expression subsequent ( 2 representing the positive feedback loop of 2 promoters and 2 promoter activators), and yellow being the activation of the hypoxia sensitive promoter due to the reduction of the DOT level below the threshold of around 20%.
You can see in the animation that initially, there is a slow then rapid fall in the DOT concentration, as it falls to very low levels, hypoxia is activated shown by the sudden appearance then disappearance of the yellow lego bricks. At that time, the rate of protein expression increases as our genetic circuit is now active. A subsequent increase in DOT levels is also observed. These set of rapid changes in DOT levels is known as "The DOT Spyke", and takes place as the fermentation process changes from one in batch to fed-batch due to the consequent addition of glycerol to maintain the cells during protein expression.
THE PLAN


 "
"





 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube