Team:DTU-Denmark/Project
From 2010.igem.org
| Home | Team | Project | Parts Submitted to the Registry | Modeling | Wet Lab | Gallery |
|---|
Contents |
Bacterial Light Switch!
Introduction
Overall Goal
The goal of our project is to enable colonies of E. coli bacteria to transition between production of two different reporter proteins. In our system, switching between states will be induced by exposing the bacteria to light. Each of the states will have a specific frequency associated with it. There are multiple potential applications for biologicals "switches" such as these, this includes the improved control of production of additives in industrial biotechnological processes.
Project Concept
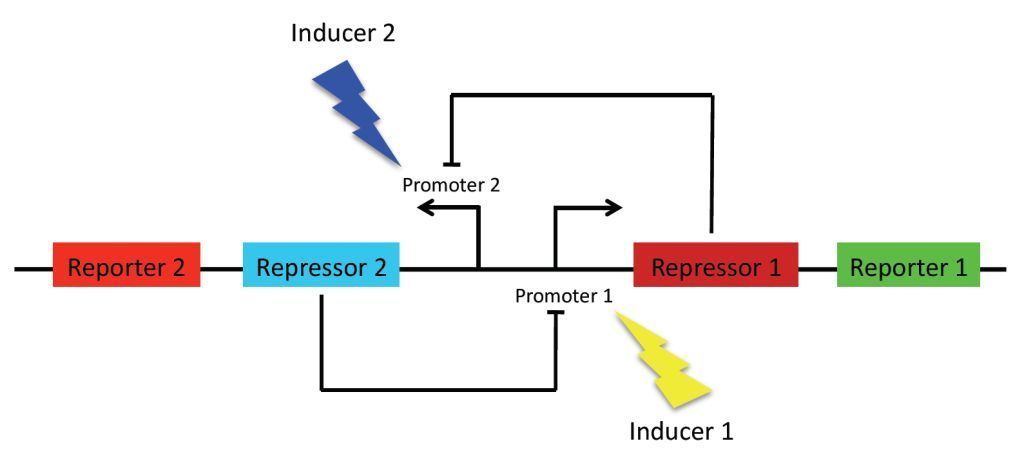
As previously stated, the main goal of our project is to design a bistable switch. The switching between the two states will be controlled by the introduction of two different wavelengths of light, each wavelength responsible for the induction of a different state. As a proof of concept, we’re using fluorescent proteins as reporter genes which makes it easy to observe and characterise the system. In principle, however, any reporter gene can be used.
Our original project concept revolved around using light-receptors to instigate the switch between the two stable state. It was thought that the production of the first reporter protein would be induced by red light (660 nm). At the same time, production of the other reporter will be suppressed by a coexpressed repressor. Conversely, production of the second reporter would be induced by blue light (470 nm). Bistability of the system is achieved by using two repressors which negatively regulate each other’s expression. This enables the system to sustain state without continuous input, i. e. once production of a reporter protein is initiated, it will persist until the system is forced into the other state.
Our project concept has since changed to using two different carbohydrate sources as a means of switching between the two stable states. This means that the state in which the bacteria will be found depends on which one of two carbohydrate sources it was last exposed.
Background
Repressors
Alpha-repressor
The C1-repressor is responsible for repressing transcription of the lytic genes, thereby maintaining the stable lysogenic state. The induction of the lytic state is caused by activated RecA, which stimulates the self-cleavage of the C1-repressor. We will be using the C1-repressor in our system.
Transcription Termination and Anti-Termination
Termination
Termination can fall into one of two catagories:
- Intrinsic Termination
- Factor-dependent Termination
Intrinsic Termination can be found to occur at defined template sequences, usually a region of hyphenated inverted sequence symmetry followed by a run of T residues. Termination through intrinsic terminators is stimulated by additional factors, e.g. NusA. Termination occurs due to the stem-loop structure formed by the base-pairing of mRNA with itself caused by inverted sequence symmetry, followed by the run of T residues. The NusA protein causes the RNA-p complex to temporarily stall at the stem-loop structure, when this is followed by a poly-A tail, the RNA-DNA duplex is destabilized. This causes the RNA-p to dissociate from the DNA, thereby terminating transcription.
Factor-dependent Termination occurs due to events that are not directly related to transcription, such as the release of ribosomes from nascent transcript or DNA damage. One such host termination factor is Rho, which acts on many sites along the bacterial chromosome. MFD, is a host termination factor that is responsible for releasing RNA-p stalled at sites of UV-induced DNA lesions.
Anti-Termination
Anti-Termination is the process by which the termination of gene transcription is prevented. Such control of gene transcription can be found in the phage Lambda system. The mechanism is controlled by proteins, such as the lambda N or lambda Q-proteins. The expression of early genes and late genes are both regulated by the anti-termination mechanism, controlled by the lambda N-protein and the lambda Q-protein, respectively. The N-protein is able to suppress transcription termination at both factor-dependent and factor-independent termination sites. N anti-termination is strongly stimulated by the NusA protein. Unlike the N-protein, the Q-protein specifically binds to a DNA sequence immediately upstream of the pR´ promoter.
A more detailed explanation of these anti-termination mechanisms will be posted later on.
N-protein plasmid
The N protein were isolated from salmonella genomic DNA with specific designed primers. We used the natural occurring RBS site, as a High expression of N have shown non specific anti-termination effect on a global scale on the genome. #References References
 "
"