Team:Stockholm/3 September 2010
From 2010.igem.org
Contents |
Andreas
Transfer of m-yCCS into pEX & Cloning of N-CPPs
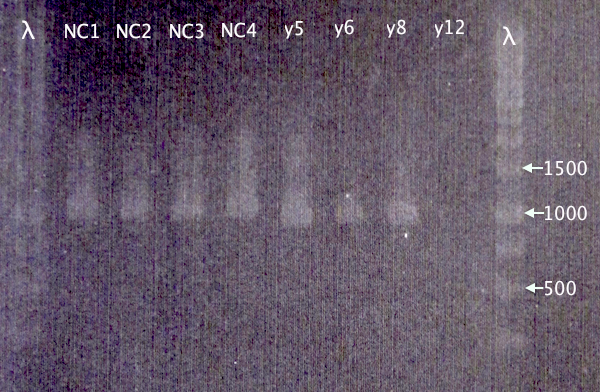
Restreak results from 2/9 revealed four white (positive) clones. These were picked for colony PCR (y5, y6, y8 and y12).
Also picked four colonies from N-CPP plate from 30/8 (NC1, NC2, NC3 and NC4).
Colony PCR
| PCR tubes | |
|---|---|
| dH2O | 16.2 |
| DreamTaq buffer | 2 |
| 10 mM dNTPs | 0.4 |
| pEXf | 0.4 |
| pEXr | 0.4 |
| DNA | 0.5 |
| DreamTaq pol. | 0.08 |
| Total | 20 μl |
Gel verification
1 % agarose, 80 V
Expected bands
- pSB1C3.N-CPPs: 365 bp (TAT), 374 bp (LMWP), 395 bp (Tra10)
- pEX.m-yCCS: 963 bp
Results
Correct bands for pEX.m-yCCS clones 5, 6 and 8. No band for y12. 5 & 8 will later be selected for plasmid prep.
Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted.
Cloning of His⋅SOD into pMA
Plasmid prep
From 2/9 ON cultures
Elution: 50 μl x2
| DNA concentrations | ||
|---|---|---|
| Sample | Conc. [ng/μl] | A260/A280 |
| pSB1C3.m-yCCS 1 | 143.4 | 1.91 |
Sequencing
pSB1C3.m-yCCS: ASB0045 A55
Glycerol stock
pMA.His⋅SOD 2010-09-03
Cloning of N-CPPs into pSB1C3
Re-ligation of 30/8 digestion.
Ligation
[Dig. pSB1C3 X+A EXTR 1] = 13.72 ng/μl [Dig. N-CPP X+A] = 66.6 ng/μl
| Ligation mix | |
|---|---|
| Vector DNA | 6 |
| Insert DNA | 9 |
| 5X Rapid Ligation buf. | 4 |
| dH2O | 0 |
| T4 DNA ligase | 1 |
| Total | 20 μl |
Transformation
Standard transformation procedures.
- 2 μl ligation mix†
- 5 μl ligation mix
†: Possibly contaminated due to non-sterile transformation.
Gel verification of digestion sample
- NCPP: undigested N-CPP cluster plasmid
- Dig NCPP: digested N-CPP cluster plasmid, XbaI and AgeI
1 % agarose, 100 V
Results
Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.
Also strange is that it looks like there are two bands present in the undigested sample.
Mimmi
Saftey issues on the home page
- Making a first layout to answer the saftey questions...
pMA.his.SOD
- Plasmid prep. following E.Z.N.A. plasmid mini kit protocol 1
- Wash two times with DNA wash buffer
- Eluate in 50µl sH2O
DNA concentration
266ng/µl
MITF-M
Colony PCR
Fermentas Pfu
| Mix | (µl) | x5 | Primers | conditions | ||||
|---|---|---|---|---|---|---|---|---|
| sH2O | 17 | 85 | pSB_VF2 | time | °C | |||
| 10x buffer | 2.5 | 12.5 | pSB_VR | 2m | 95 | |||
| dNTP | 2.5 | 12.5 | 30s | 95 | ) | |||
| F primer | 1 | 5 | 30s | 55 | > 30 cycles | |||
| R primer | 1 | 5 | 2m40s | 72 | ) | |||
| DNA | 0.5 | 5x0.5 | 10m | 72 | ||||
| Pfu pol | 0.5 | 2.5 | oo | 10 | ||||
| tot | 25µl | 125µl | ||||||
pMA.his.SOD
Digestion
| Mix | (µl) | (µl) | Conditions | [pMA.SOD.his] = 246ng/µl -> ~2µg = 8µl | |||
|---|---|---|---|---|---|---|---|
| DNA | 8 | 7.5 | Time | °C | [pMA.his.SOD] = 280ng/µl -> ~2µg = 7.5µl | ||
| s2O | 17 | 17.5 | 30m | 37 | [pSB1C3] = 148ng/µl -> ~1.2µg = 8µl | ||
| 10x buffer | 3 | 3 | 20m | 65 | |||
| XbaI | 1 | 1 | |||||
| PstI | 1 | 1 | |||||
| tot | 3x30µl | ||||||
Gel
| well | sample |
|---|---|
| 1 | 1kb ladder |
| 2 | pMA.SOD.his |
| 3 | pMA.his.SOD |
| 4 | pSB1C3.RFP |
- Cut gel bands
- Save in fridge
 "
"