Team:Freiburg Bioware/testpage
From 2010.igem.org

18. Labortag 01.06.2010: Modifying MCS of pAAV_MCS vector
Investigators: Anissa, Adrian, Bea, Chris W., Hanna, Patrick, Volker, Sven
Oligos received from Sigma-Aldrich
(right ITR of pAAV_MCS, left ITR of pAAV_MCS and MCS RFC25 for pAAV)
- Hybrization of received oligos: MCS RFC25 for pAAV (forward) and MCS RFC25 for pAAV (reverse)
- Centrifuge tubes prior to open tubes (13.000 rpm, 30 sec)
- MCS RFC25 for pAAV (forward): Add 92µL Millipore H2</sup>O (Volume on obtained sheet)
- MCS RFC25 for pAAV (reverse): Add 394 µL Millipore H2</sup>O (Volume on obtained sheet)
- Vortex the resuspended DNA
- Make aliquots of both Oligos (1:10): 10 µL Oligo + 90 µLH2</sup>O (final volume usually 100 µl)
- Mix together(into PCR-tube):
| Volume/µL | solution |
| 10 (1:10) | Oligo 1: MCS RFC25 for pAAV (forward) |
| 10 | Oligo 2: MCS RFC25 for pAAV (reverse) |
| 4 | 100mM TrisHCl pH8 |
| 8 | 5mM MgCl2 |
| 8 | H20 |
- Program: ORIGAMI 1 modified for long oligos:
- 1 99°C 7’
- 2 99°C 1’
- -1°C R=0.3 °/s
- Goto 2 rep 74
- Hold 4°C
- While hybridization of oligos is performed, digestion of pAAV_MCS vector can be conducted
following standard protocol for cloning.
- Title: Ligation MCS_Oligo with pAAV_MCS
- Plasmid: pAAV_MCS
- Buffer used: 3
- BSA: Yes
- Measure DNA-concentration with Nanodrop
- DNA-Concentration:260 ng/uL
- Restriction-enzyms used: http://www.neb.com/nebecomm/DoubleDigestCalculator.asp
Enzyme1 (Nr. Lab: 152): ClaI
Enzyme2 (Nr. Lab: 15): BglII
- Digestion components :
| components | pAAV_MCS |
| DNA | 4 |
| BSA (10x) | 3 |
| Buffer 3 (10x) | 3 |
| Enzyme: ClaI (no.Lab:152) | 2 |
| Enzyme: BglII (no.Lab:15) | 1 |
| H2</sup>O | 17 |
| Total volume | 30 |
- Incubate for 1,5 h at 37°C
- 1% Agarose gel
- 1% agarose gel was prepared, gel ran for 45 minutes( first: 90V, after 15 minutes: 115 V)
- Amount of loading dye added
| sample/µL | loading dye/µL |
| marker: 8 | contains loading dye |
| pAAV_MCS: 24 | 6 |
- Expected size of fragments
| sample | expected size |
| pAAV_MCS: cut with ClaI and BglII | 4580 bp |
19. Labortag 02.06.2010: Oligos (NotI)
Investigators: Adrian, Bea, Chris W., Hanna, Anissa
Practical work:
Control plate contained no clones. :)
4 colonies were picked and grown @ 37°C over night.
Theoretical work:
Oligos for site directed mutagenesis of the NotI restriction sites in pAAV_MCS (ITRs) were designed:File:Freiburg10 NotI ITR Oligos.pdf
Sponsoring work:
Sponsoring letter was adapted for Quiagen.
20. Labortag 03.06.2010: pAAV_RFC25_MCS -> problem
Investigators: Anissa, Bea, Melanie, Christian L.
Comment: Continue with Mini-Prep and test digestion of pAAV_RFC25_MCS
Mini-Prep and test digestion have been performed:
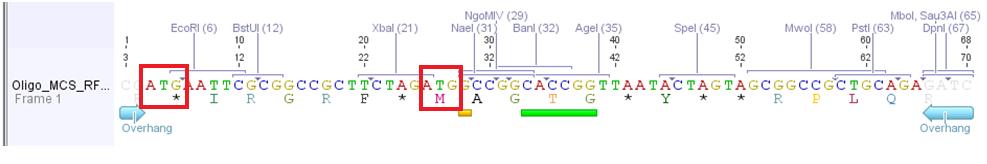
Problem: Designed oligos (MCS_RFC25) for altering the MCS of the pAAV_MCS vector cannot be used.
Two startcodons are in the MCS.
The two startcodons are not in the same open reading frame (ORF). Therefore two proteins will be produced. The Gene of Interest and the short peptide (30 aa).
The idea of the oligos was to ligate the oligos into the pAAV_MCS vector. The oligos contained two overhangs which correspond to the sequences of the two restriction sites ClaI and BglII:
The pAAV_MCS vector was digested with ClaI and BglII and then we ligated the oligo and the vector. Problem was that we did not notice that the overhang of ClaI and the sequence of EcoRI of the MCS_RFC25 resulted in another ATG startcodon.
Possible Solutions:
- first: modify MCS with ordered oligos of Sven (shorter MCS which cannot be used for pEX)and clone mVenus
- Perform site-directed-mutagenesis (QuikChange from Stratagene)
- Order new MCS-oligos and consider that no new ATG is produced. For example: add another base between ClaI overhang and EcoRI sequence. -----ATXG---- This solution is the more possible one we are going to perform.
Practical work
- Preparing four glycerol stocks (2:1)
- numbers: B4 - B7 (for details see nomenclature)
- stored in -80°C, Box 1
- MiniPrep
- Nanodrop concentrations
| Sample | Concentration/ng*µl-1 |
| P11 | 340,5 |
| P12 | 364,0 |
| P13 | 358,5 |
| P14 | 284,4 |
- Test digestion
| Components | Volume µl | Mastermix µl |
| DNA | 800 | -- |
| BSA (10x) | 1,5 | 7,5 |
| Buffer No.2 (10x) | 1,5 | 7,5 |
| Enzyme 1 (no.Lab:45) Nde I | 0,5 | 2,5 |
| Enzyme 2 (no.Lab:71) Spe I | 0,5 | 2,5 |
| H2</sup>O | variable | -- |
| Total volume | 15 | 20 |
| Sample | Volume/ µl | H2</sup>O / µl |
| P11 | 2,3 | 8,7 |
| P12 | 2,2 | 8,8 |
| P13 | 2,2 | 8,8 |
| P14 | 2,8 | 8,2 |
- Incubation: 1,5 h
- Materials
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P11 | 15 µl | 3 µl | 3677 bp | 974 bp |
| P12 | 15 µl | 3 µl | 3677 bp | 974 bp |
| P13 | 15 µl | 4 µl | 3677 bp | 974 bp |
| P14 | 15 µl | 4 µl | 3677 bp | 974 bp |
| Marker | Sample P11 /18 µl | Sample P12 /18 µl | Sample P13 /19 µl | Sample P14 /19 µl | |
|---|---|---|---|---|---|
| Lane | 1 | 3 | 5 | 7 | 9 |
Results of agarose-gel:
- Expected fragments of 3677 bp and 974 bp can be cerified. The insertion of the RFC25_MCS has been inserted.
Picking clones of Thymindinkinase of Amor
- 5 clones of the XL-1 Blue colonies containing the plasmid pUB6/HV5/His6 with the thymidinkinase have been picked from LBamp-agarplates
- 1 clone of the XL-1 Blue colonies containing the plasmid pUB6/HV5/His6 without the thymidinkinase (control) has been picked from LBamp-agarplates
- all clones have been inoculated in 10 mL LB containing 10 µL Amp. Incubation: 37°C over-night.
- to do: Mini-Prep of pUB6/HV5/His6 with the thymidinkinase and pUB6/HV5/His6 without the thymidinkinase (control)
Idea
- Insertion of Kozak consensus sequence before MCS to enhance gene expression (cloning consideration in Stratagene manual)
- New RFC standard with Kozak sequence for eucaryotes??
- pXX6 alle Adenovirus Helfer Gene ohne AAV Gene; Plasmid von J. Samulski; evtl. Anfragen für Benutzererlaubnis
- pKEX-2XL.Rep40 Expressionsplasmide für das Rep Proteine 40
- pKEX-2XL.Rep 52 Expressionsplasmide für das Rep Proteine 52
- pKEX-2XL.Rep 68 Expressionsplasmide für das Rep Proteine 68
- pKEX-2XL.Rep 78 Expressionsplasmide für das Rep Proteine 78
- pCMV-VP(HS) Expressionsplasmid der drei VP Proteine in Kombination (assemly kompetent
- pKEX-VP1 Expressionsplasmide für die einzelnen VP (cap) Proteine (VP1, VP2 und VP3) vorsicht: sind alleine nicht assembly kompetent; Ruffing et al 1992; Steinbach et al. 1997;
- pKEX-VP2Expressionsplasmide für die einzelnen VP (cap) Proteine (VP1, VP2 und VP3) vorsicht: sind alleine nicht assembly kompetent; Ruffing et al 1992; Steinbach et al. 1997;
- pKEX-VP3Expressionsplasmide für die einzelnen VP (cap) Proteine (VP1, VP2 und VP3) vorsicht: sind alleine nicht assembly kompetent; Ruffing et al 1992; Steinbach et al. 1997;
- pTRUF_CMV_eGFPEinzelstrang Vektor zur Expression von eGFP
- dsAAV_CMV_eGFP"self complementary" Expressionsvektor von X. Xiao; evtl. anfragen wegen Benutzererlaubnis
- pTAV2 (gesamtes AAV Genom im "blue script" Vektor)
- pDG komplettes Helferplasmid zur Vektorherstellung; Grimm et al. 1998
- In order to obtain the DNA following steps have been performed:
- cut out the spot where the DNA is spotted with a clean scalpel (note: scalpel should not have any contamination).
- put a 0,5 mL Eppi in a 1,5 mL Eppi. Put little holes in the smaller eppi.
- transfer Whatman paper into Eppi
- add 50 µL TE-EF (Redissolving buffer) to whatman paper and wait 15 minutes
- centrifuge eppis at 2000 rpm, 10 minutes
- Transformation with obtained plasmids was performed.
21. Labortag 04.06.2010: DKFZ plasmid Retrafos, TK/GMK Mini-Prep
Investigators: Adrian, Bea, Chris W., Hanna
DKFZ
Comments: Plasmids of PD Kleinschmidt of the DKFZ arrived. The DNA was dried on a whatman paper.
Plasmids received:
Fusion Enzyme: Thymidinkinase/Guanylate Kinase (TK/GMK)
Plasmid Mini-Prep
- experiment date: 04.06.2010 ; time: 3,5h
- name of investigator: Adrian, Bea, ChrisW.,Hanna
- new vector name: pUB_V5_His6 + TK/GMK (fusionenzyme)
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Control | |
| Bacteria strain | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue |
| Plasmidname | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK |
| Date | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 |
| given number | B8 | B9 | B10 | B11 | B12 | B13 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Control | |
| given number | P15 | P16 | P17 | P18 | P19 | P20 |
Nanodrop concentration
- Plasmid
- Given Plasmid-Number: P15; DNA concentration: 493,7 ng/µL ;
- Given Plasmid-Number: P16; DNA concentration: 464,4 ng/µL;
- Given Plasmid-Number: P17; DNA concentration: 445,6 ng/µL;
- Given Plasmid-Number: P18; DNA concentration: 562,9 ng/µL;
- Given Plasmid-Number: P19; DNA concentration: 528,1 ng/µL;
- Given Plasmid-Number: P20; DNA concentration: 499,9 ng/µL;
Comments:A Plasmid-Mini Prep with the received Fusionenzyme Thymidinkinase/Guanylate Kinase (TK/GMK)from Amor has been performed.
The DNA will be sent to GATC for sequencing.
- pUB6_V5_His6 - clone 1 (P15) (3 µL added to 27µL H20) has been sent to GATC.
- Expected results: Saturday
22. Labortag 05.06.2010: Insertion of iGEM expression parts into pAAV_MCS
Investigators: Adrian, Bea, Melanie, Hanna
Hybridization:
- Hybridization of received oligos: iGEM expression parts (RFC25 without EcoRI, NotI)
- Prior to opening the tubes, they were centrifugated at 13.000 rpm for 30 sec.
- expression part MCS_for (charge-no: ST00114065): 108 µL Millipore H2</sup>O (Volume on obtained sheet)were added.
- expression part MCS_for (charge-no: ST00114066): 165 µL Millipore H2</sup>O (Volume on obtained sheet)were added.
- Resuspended DNA was vortexted.
- Aliquots of both Oligos (1:10) were prepared: 10 µL Oligo + 90 µL H2</sup>O (final volume usually 100 µl).
- Mix together(into PCR-tube):
| Volume/µL | solution |
| 10 (1:10) | Oligo 1: expression part MCS_for (charge-no: ST00114065) |
| 10 (1:10) | Oligo 2: expression part MCS_for (charge-no: ST00114066) |
| 4 | 100 mM TrisHCl pH8 |
| 8 | 5 mM MgCl2 |
| 8 | H20 |
- Program: ORIGAMI 1 modified for long oligos:
- 1 99°C 7’
- 2 99°C 1’
- -1°C R=0.3 °/s
- Goto 2 rep 74
- Hold 4°C
- While hybridization of oligos was performed, digestion of pAAV_MCS vector was conducted
following standard protocol for cloning.
Digestion:
- Title: Ligation iGEM expression parts (="iGEM-MCS") with pAAV_MCS
- Plasmid: pAAV_MCS
- Buffer used: 3
- BSA: Yes
- DNA-Concentration: 260 ng/uL
- Restriction-enzyms used:
Enzyme1 (Nr. Lab: 152): ClaI
Enzyme2 (Nr. Lab: 15): BglII
- Digestion components :
| components | pAAV_MCS |
| DNA | 5.8 µL |
| BSA (10x) | 3 µL |
| Buffer 3 (10x) | 2 µL |
| Enzyme: ClaI (no.Lab:152) | 2 µL |
| Enzyme: BglII (no.Lab:15) | 1 µL |
| H2</sup>O | 16.2 µL |
| Total volume | 30 µL |
- Mixture was incubated for 1,5 h at 37°C.
Agarose-Gel:
- 1% agarose gel was prepared, gel ran for 45 minutes(110 V)
- Amount of loading dye added
| sample/µL | loading dye/µL |
| marker: 8 | contains loading dye |
| pAAV_MCS: 30 | 6 (6x loading dye) |
- Expected size of fragments
| sample | expected size |
| pAAV_MCS: cut with ClaI and BglII | 4580 bp |
</ul>
Gelextraction:
Gel measurement:
| Sample | weight |
| pAAV_MCS | 60 mg |
- Gelextraction was performed following standard protocol.
- DNA-concentrations were meassured: pAAV_MCS 18.2 ng/µL, Oligos: 136.7 ng/µL -> 1:10 dilution was prepared: 13.67 ng/µL
Ligation:
| iGEM-MCS | pAAV_MCS | |
| Volume/µl | 0.4 | 8.6 |
Trafo was performed (using XL1B cells) following standard protocol.
23. Labortag 07.06.2010: Mini-Preps (Kleinschmidt-plasmids and pAAV_iGEM-MCS)
investigators: Achim, Kira,Jessy, Chris W., Hanna, Adrian, Bea
pAAV_iGEM-MCS
Plasmid Mini-Prep
- experiment date:07.06.2010; time: whole day
- name of investigator: Achim, Kira,Jessy, Chris W., Hanna, Adrian, Bea
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| Bacteria strain | XL1B | XL1B | XL1B | XL1B |
| Plasmidname | pAAV_iGEM-MCS | pAAV_iGEM-MCS | pAAV_iGEM-MCS | pAAV_iGEM-MCS |
| Date | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 |
| given number | - | B27 | B28 | B29 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| given number | - | P34.2 | P34.3 | P34.4 |
Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) AgeI ; Enzyme 2 (no.Lab:___) NdeI
- Plasmid
- Given Plasmid-Number: P34.2; DNA concentration: 433.78 ng/µL ;
- Given Plasmid-Number: P34.3; DNA concentration: 408.48 ng/µL ;
- Given Plasmid-Number: P34.4; DNA concentration: 409.80 ng/µL ;
Comments:Clone no.1 was dismissed...
Test digestion:
| Components | Volume/µL | Mastermix |
| DNA (clone 2) | 1000 ng | - |
| BSA (10x) | no | - |
| Buffer no. 4 (10x) | 1.5 µL | - |
| Enzyme 1 (no. Lab: ) AgeI | 0.75 µL | - |
| Enzyme 2 (no. Lab: ) NdeI | 0.5 µL | - |
| H2</sup>O | variable | - |
| Total volume | 15 µL | - |
| Sample | Volume sample/ µl | Volume H2</sup>O / µl |
| P34.2 | 2.3 | 9.95 |
| P34.3 | 2.4 | 9.85 |
| P34.4 | 2.4 | 9.85 |
- Incubation: 45 min, 37°C
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1%), 3 µL GELRED (3-6µl), at 110 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 15 µl | 3 µl | 3686 bp | 951 bp |
| P34.3 | 15 µl | 3 µl | 3686 bp | 951 bp |
| P34.4 | 15 µl | 3 µl | 3686 bp | 951 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample | Sample | Sample | |
|---|---|---|---|---|
| Lane | 34.2 / 18 µl | 34.3 / 18 µl | 34.4 / 18 µl |
Kleinschmidt-plasmids
Plasmid Mini-Prep
- experiment date: 07.06.2010 ; time: 10 – 20 h
- name of investigator: Kira, Achim, Jessy, Bea, Adrian, Hanna
- Kleinschmidt-plasmids
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | Clone 9 | Clone 10 | Clone 11 | Clone 12 | Clone 13 | |
| Bacteria strain | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B |
| Plasmidname | pXX6 | pKEX-2XL.Rep 40 | pKEX-2XL.Rep 52 | pKEX-2XL.Rep 68 | pKEX-2XL.Rep 78 | pCMV-VP(HS) | pKEX-VP1 | pKEX-VP2 | pKEX-VP3 | pTRUF_CMV_eGFP | dsAAV_CMV_eGFP | pTAV2 | pDG |
| Date | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 |
| given number | B14 | B15 | B16 | B17 | B18 | B19 | B20 | B21 | B22 | B23 | B24 | B25 | B26 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | Clone 9 | Clone 10 | Clone 11 | Clone 12 | Clone 13 | |
| given number | P21 | P22 | P23 | P24 | P25 | P26 | P27 | P28 | P29 | P30 | P31 | P32 | P33 |
| measured concentration | 351,01 | 673,1 | 532,22 | 579,05 | 725,31 | 659,68 | 692,8 | 545,46 | 568,34 | 420,62 | 446,95 | 496,8 | 472,58 |
Comments:
Many things went wrong today!
- Glycerol stocks must be vortexted!
- Check mini-prep buffers - especially buffer PE (needs to contain ethanol!)
- Always check volumes - try to estimate if volume makes sense (check pipettes!)
- Don't discard bacteria cultures until glycerol stocks and mini-preps are successfully done!!!
Today's conclusion: Better ask 2 times than do something wrong without asking!!!
Site-directed mutagenesis of pAAV_iGEM-MCS (PstI)
Quickchange site directed mutagenesis:
PCR reaction:
- 2.5 µL 10x Pfu Ultra II buffer
- 0.5 µL template (therefore the a 1:20 dilution of the pAAV_iGEM-MCS (433 ng/µL) was prepared) = 10.825 ng
- 0.56 µL primer 1 (of 1:10 dilution)
- 0.56 µL primer 2 (of 1:10 dilution)
- 1 µL DMSO (primers form very strong secondary structures)
- 0.5 µL dNTP
- 18.88 µL dH2</sup>O
- 0.5 µL PfuUltra II fusion (1.25 U)
-> end volume: 25 µL
PCR program:
1 x : 2' 95°C (HotStart polymerase)
20 x : 30 s 95°C -> 1' 55°C -> 5' 68°C
1 x : 4°C (over night)
Experiment will be continued tomorrow.
24. Labortag 08.06.2010: Cloning of mVenus_YFP into pAAV_iGEM-MCS, continuation of site-directed mutagenesis
Investigators: Kira, Jessy, Hanna, Achim
Analysis of iGEM-MCS sequence (RFC25 without EcoRI and NotI):
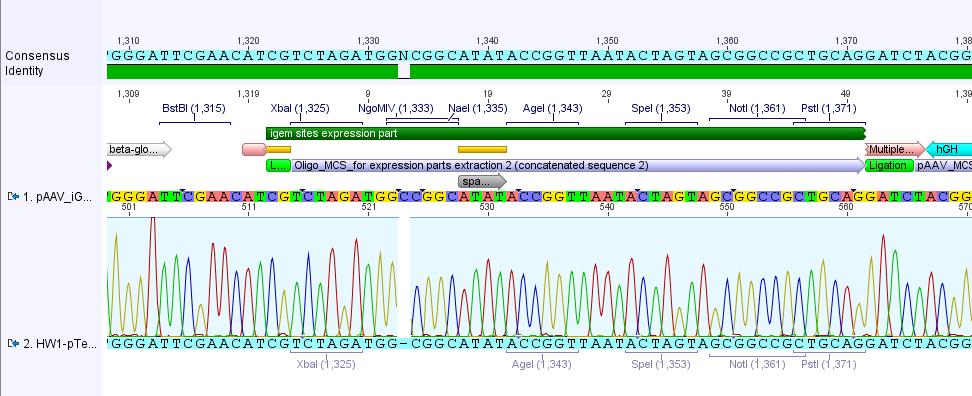
The alignment with the theoretical pAAV_iGEM-MCS delivered a "C-deletion" within the NgoMIV restriction site. Due to that the pAAV_iGEM-MCS lacks this site. We controlled the bill of delivery and noted that the deletion must be GATC's fault.
All in all this doesn't matter, because parts which are in iGEM standard can be inserted and therefore they will deliver the lacking NgoMIV restriction site. Further on this could be an advantage because after digestion the fragment which is cut out can be better detected in the gel.
Insertion of mVenus_YFP into pAAV_iGEM-MCS
Digestion:
| components | V (pAAV_iGEM-MCS)/ µl | I(pGA14mVenus_Geneart) / µl |
| DNA | 2,3 | 3,8 |
| BSA (10x) | 2 | 2 |
| Buffer 3 (10x) | 2 | 2 |
| Enzyme: AgeI (no.Lab:149) | 1,25 | 1,25 |
| Enzyme: XbaI (no.Lab:63) | 0,75 | 0,75 |
| H2</sup>O | 11,7 | 10,2 |
| Total volume | 20 | 20 |
0.5 g Agarose, 50 ml TAE (1%), 3 µL GELRED (3-6µl), at 115 Volt, running time:
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 20 µl | 4 µl | 4617 bp | 22 bp |
| pGA14mVenus | 20 µl | 4 µl | 2870 bp | 774 bp |
| Sample | weight | concentration |
| Vektor_Oben | 0,32 g | 29,8 ng/µl |
| Vektor_Unten | 0,14 g | 12,8 ng/µl |
| Insert | 0,3 g | 12,9 ng/µl |
- Vector: 4,16 µl
- Insert: 4,84 µl
- Vector: 6 µl
- Insert: 3 µl
continuation of site-directed mutagenesis
Digestion with DpnI:
Trafo:
25. Labortag 09.06.2010: pAAV_iGEM-MCS_mVenus-YFP, site-directed mutagenesis
investigators: Jessy, Achim, Sven, Toby, Hanna
No pAAV_iGEM-MCS_w/oPstI transformed bacteria (site-directed mutagensis) grew on the ampicillin agar plates over night!
Experiment is conducted again by Toby (Thanks a lot!!!).
Further on the cloning of mVenus-YFP into pAAV_iGEM-MCS seemed to fail:
Only on the agar plate cotaining the bacteria transformed with the "vector_oben" ligation, which actually should be the "wrong" ligation (gelextraction of a band which contained fragments that were too large, see picture in lab journal), revealed colonies. Therefore this experiments is also conducted one more time by Sven (thanks also a lot!!!)
Insertion of mVenus_YFP into pAAV_iGEM-MCS
- experiment date: 9.6.2010
- name of investigator: Sven
- plasmid:
- Vector: name: pAAV_iGEM-MCS number: 34.2 production date: 5.6.2010
- Insert: name: pOG14_mVenus number: - production date: ____ origin: Sven :)
- new vector name: pAAV_iGEM-MCS_mVenus-YFP
- buffer used: NEB4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) AgeI ; Enzyme 2 (no.Lab:___) XbaI
- DNA concentration (vector): 433 ng/µL ; DNA concentration (insert): 530 ng/µL
Digestion
| components | volume of vector /µl | volume of insert /µl |
| DNA | 2.83 | 2.77 |
| BSA (100x) | 0.5 | 0.5 |
| Buffer NEB4 (10x) | 3.0 | 3.0 |
| Enzyme 1 (AgeI) | 1.75 | 1.75 |
| Enzyme 2 (XbaI) | 1.25 | 1.25 |
| H2</sup>O | 20.73 | 20.67 |
| Total volume (e.g. 15,20,25,30 µl) | 30 | 30 |
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED (3-6µl), at 115 Volt
Gelextraction
- insert (mVenus-YFP): 26 ng/µL
- vector (pAAV_iGEM-MCS): 30 ng/µL
Ligation
- vector: 5.85 µL
- insert: 3.15 µL
- ligase: 1 µL
- buffer: 10 µL
Test digestion of pAAV_iGEM-MCS
p style="font-size:15px; font-weight: bold; color: blue;">Test digestion</p>
- buffer used: 1 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) NgoMIV
- Plasmid
- Given Plasmid-Number: P34.2 ; DNA concentration: 433.78 ng/µL;
- Given Plasmid-Number: P34.3 ; DNA concentration: 408.48 ng/µL;
- Given Plasmid-Number: P34.4 ; DNA concentration: 409.8 ng/µL;
| Components | P34.2 [µL] | P34.3 [µL] | P34.4 [µL] |
| DNA | 2.3 | 2.5 | 2.4 |
| BSA (10x) | 1 | 1 | 1 |
| Buffer no. 1 (10x) | 1 | 1 | 1 |
| Enzyme 1 (no. Lab: 113) ngoMIV | 1 | 1 | 1 |
| H2</sup>O | 4.7 | 4.5 | 4.6 |
| Total volume | 10 | 10 | 10 |
- Incubation: 1 h, 37°C
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes + 20 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 10 µl | 2 µl | 3728 bp | 903 bp |
| P34.3 | 10 µl | 2 µl | 3782 bp | 903 bp |
| P34.4 | 10 µl | 2 µl | 3782 bp | 903 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample P34.2 /µl | Sample P34.3 /µl | Sample P34.4 /µl | |
|---|---|---|---|---|
| Lane | 8 µL | 10 µL | 10 µL | 10 µL |
Comments: Digestion of P34.2 delivered just one fragment size, revealing that - as expected after sequencing (C-deletion: no NgoMIV site in iGEM-MCS) - there was just one NgoMIV restriction site in the vector. In contrast to that the digestion of P34.3 and P34.4 delivered two bands - whereas the band of the smaller fragments looked like a smier. Because of this obscure and non-satisfying results (besides our assumption that there's something wrong with the marker :) )we decided to sequence the P34.3 vector:
- c(P34.3): 408.48 ng/µL
- Volume (plasmid): 5.14 µL
- Volume (vector): 24.86 µL
- Name of eppi: HW_34
- Primer: GATC_std_pTeSp-1
Picking of Clones from pAAV_iGEM-MCS_mVenus-YFP_Vektor_oben
- experiment date: 9.6.2010
- name of investigator: Hanna, Achim
- plasmid:
- Vector: name: pAAV_iGEM-MCS_Vektor_oben
- Insert: name: pOG14_mVenus
- new vector name: pAAV_iGEM-MCS_mVenus-YFP_Vektor_oben
The Clones were picked and incubated in 10 mL LB-Medium containing 10µL ampicillin over night at 37°C on a rotary shaker.
 "
"