Team:Stockholm/9 August 2010
From 2010.igem.org
Contents |
Hassan
Folate
below links and articles are meant to be study when I'm finished with interaction map and prove of concept!
- [http://www.ncbi.nlm.nih.gov/pubmed/12761857]
- [http://www.ncbi.nlm.nih.gov/pubmed/17307853,18403248,17698004,17220358,16359333,10201405,15489424,2254281,15865426,17923103,11041843,2185835,11371182,9287033,1958664,8208138,8019142,8408018,8634297,1098865?report=docsum]
- [http://www.ncbi.nlm.nih.gov/pubmed/15611104]
- [http://www.ncbi.nlm.nih.gov/pubmed/17698004]
- [http://www.ncbi.nlm.nih.gov/pubmed/8208138]
- [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide]
Possible Genes
- decrease in CAT (Catalase) : [http://www.ncbi.nlm.nih.gov/pubmed/10537016][http://onlinelibrary.wiley.com/doi/10.1034/j.1600-0749.2003.00087.x/abstract]
- increase in SOD (SOD3) : [http://www.ncbi.nlm.nih.gov/pubmed/12692376]
- possible genes involved in Melanocytes adhesion: "An imbalance in cell adhesion molecules was also observed in vitiligo melanocytes. Differentially downregulated genes in vitiligo melanocytes included integrin alpha 7, integrin beta-like 1, integrin beta 7 and catenin alpha-like 1 (cadherin-associated protein). The adhesion between the melanocytes and the basal membrane is mediated by integrins and the contacts between melanocytes and keratinocytes are mediated by cadherins (Hara et al., 1994; Tang et al., 1994; Zambruno et al., 1993). The importance of regulated expression levels of cadherins have also been shown during migration of melanoblasts (Nishimura et al., 1999). It is difficult to predict if this specific downregulation of adhesion molecules impairs the attachment of the melanocytes in the basal layer of the epidermis; however, detachment and transepidermal loss of melanocytes have previously been reported in vitiligo vulgaris (Gauthier et al., 2003)."[http://www.ncbi.nlm.nih.gov/pubmed/18426409] May also look at [http://www.ncbi.nlm.nih.gov/pubmed/17850512] if more interested.
And those from July 22 with data from planning will bring enough explanation for interaction map.
I think by end of this week a wall of text will appear in modelling page!
Nina
Overnight culture of Tyrosinase
I inoculated eight falcon tubes containing 12 ml LB and 24 ul amphicillin (50mg/ml) with the tyrsoinase site directed mutagenesis colonies number 1, 2, 3, 4, 5, 6, 7 & 8. These were incubated overnight in 37 °C with shake and will be miniprepped tomorrow.
Digestion of protein A (ZZ domain) and bank vector C
I cut a previously PCR product of protein A ZZ domain. The first digestion was carried out with the restriction enzyme NgoMIV. However since this enzyme is from New England BioLabs and has another buffer composition compared to the second digestion with the restriction enzyme AgeI from Fermentas (fast digest) I had to run a gel clean up before following up with the second digest after the first digest.
I also cut IgG protease in the bank vector C since I wanted to replace the insert of IgG protease in bank vector C with an insert of protein A ZZ domain.
First digest:
- DNA vector with IgG protease 20 ul
- Restriction enzyme NgoMIV 1 ul
- Buffer #4 3 ul
- H2O 6 ul
- DNA PCR product of protein A ZZ domain 10 ul
- Restriction enzyme NgoMIV 1 ul
- Buffer #4 3 ul
- H2O 6 ul
I incubated the vector in 3 hours and the PCR product in 1 hour, both in 37 °C.
After 3 hours of incubation of the vector I added 1.5 ul CIAP to this sample in order to dephosphorylate the vector to prevent the vector to couple back into a circle. The sample was yet again incubated in 37 °C for 30 min.
Gel clean up of digest samples
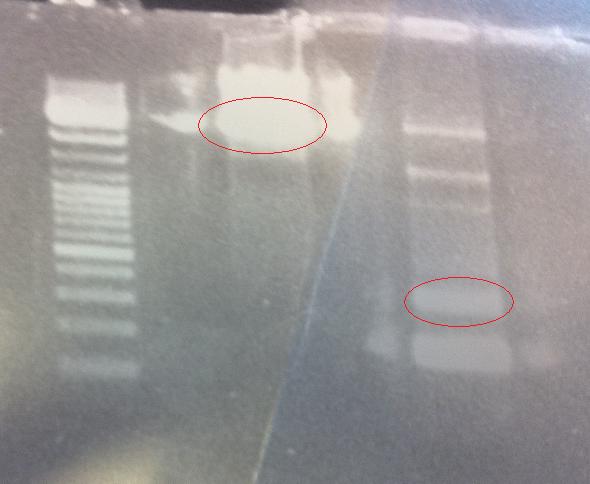
I ran the digested samples PCR product protein A ZZ domain and IgG protease in bank vector C in an 1 % agarose gel 80 V.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gels:
I cut out the red marked bands with a scalpel and carried out a gel clean up according to the method decribed in protocols.
Gel clean up:
- Protein A ZZ domain: 300 mg * 3 = 900 ul
- IgG protease: 400 mg * 3 = 1200 ul
Mimmi
MITF
- Gel
- No products!
- --> Trying more cycles of low annealing temperature
MITF + yCCS and RFP
Verification PCR
| Mix | (µl) | X5 | X3 | yCCS/RFP | MITF | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| sH2O | 22.5 | 112.5 | 67.5 | pSB1_VF2 | MITF_F | |||||
| F primer | 1 | 5 | 3 | pSB1_VR | MITF_R | |||||
| R primer | 1 | 5 | 3 | 970/925bp | ~1300bp | |||||
| DNA | 0.5 | 4X0.5 | 2X0.5 | |||||||
| tot | 25 | conditions | conditions | |||||||
| time | °C | time | °C | |||||||
| 2m | 94 | 2m | 94 | |||||||
| 30s | 94 | ) | 30s | 94 | ) | |||||
| 30s | 60 | > 30 cycles | 30s | 60 | > 30 cycles | |||||
| 2m | 72 | ) | 2m40s | 72 | ) | |||||
| 10m | 72 | 10m | 72 | |||||||
| oo | 10 | oo | 10 | |||||||
Andreas
Vector preparation for CPP cloning
pSB1C3 vectors need to be prepared for cloning of our CPPs.
- Digested with NgoMIV (N) & SpeI (S) (for C-CPPs)
- Digested with XbaI (X) & AgeI (A) (for N-CPPs)
Digestion
Vector: pSB1C3.BBa_J18930 A (133.5 ng/μl)
| [μl] | N+S | X+A |
|---|---|---|
| 10X FD buffer | 3 | 3 |
| Vector DNA | 25 | 25 |
| dH2O | 0 | 0 |
| FD XbaI | -- | 0.5 |
| FD AgeI | -- | 0.5 |
| FD SpeI | 1† | -- |
| NgoMIV (NEB) | 1 | -- |
| 30 | 30 |
† Added after 1:30 of incubation with NgoMIV, since the latter is not a FD enzyme.
Gel verification
1 % agarose, 110 V, 25 min
Expected bands: 2070 bp (pSB1C3), 714 bp (BBa_J18930)
Results
Good digestion, no uncut plasmid. Bands with correct sizes.
Gel extraction
1 % agarose, 70 V, 1 h
| DNA concentrations | ||
|---|---|---|
| Sample | Conc. [ng/μl] | A260/A280 |
| pSB1C3 X+A 1 | 13.72 | 2.04 |
| pSB1C3 X+A 2 | 6.397 | 2.31 |
| pSB1C3 N+S 1 | 12.22 | 2.09 |
| pSB1C3 N+S 2 | 5.807 | 2.46 |
Loaded 30 μl of each sample (X+A and N+S). After electrophoresis, 260 mg samples containing digested vectors were excised. DNA purified using the E.Z.N.A. gel extraction kit, following the procedures provided. Each sample eluted twice, as follows:
- pSB1C3 X+A 1: 40 μl
- pSB1C3 X+A 2: 30 μl
- pSB1C3 N+S 1: 40 μl
- pSB1C3 N+S 2: 30 μl
Amp plate preparation
Prepared 20X Amp 100.
Cloning of IgG protease
Re-transformed Top10 with the pSB1C3.IgGp ligation mixture from 3/8 (standard transformation protocol). Plated on Cm 25, incubation ON in 37 °C.
yCCS clone analysis
Ran a gel for Mimmi, where she/we investigated my yCCS clones from 15/7, prior to mutagenesis, comparing them to the RFP (BBa_J18932) gene.
Gel verification
1 % agarose, 100 V, 30 min.
Expected bands:
- yCCS (undig): 1076 bp
- yCCS (dig): 123 bp, 257 bp, 704 bp
- RFP (undig): 1036 bp
- RFP (dig): 1076 bp
Results
yCCS B was identified as RFP, which explains the failed yCCS sequencing results. However, yCCS A seems correct, so this clone/sample will be chosen for new site-directed mutagenesis (EcoRI site removal).
 "
"