Team:KAIST-Korea/Project/References
From 2010.igem.org
Data sourcesDuring our project, we needed many data. We used data of protein-protein interactions for signal transduction pathway deciding, protein sequence and domain description for modified FGFR design, gene sequence for modified FGFR sequence synthesis, and structures or sequences of antibodies and FGF binding domain of FGFR for structural alignment. These data were acquired from Uniprot, PID, NCBI and RCSB PDB. UniprotUniprot provides sequences of many proteins and domain description of well-researched proteins. Key advantage of using Uniprot is domain description: to design engineered protein for detecting Mycobacterium without knowledge of function of each part of protein. Tuberculosis antigen MPT51 is impossible one. Uniprot provided the information of location of FGF binding domain of FGFR, which is replaced by our single-chain antibody 16A1. Uniprot also provided sequences of some antibodies to make single chain antibody sequences.
PIDPID (Pathway Interaction Database) provides the interaction networks between protein and protein or protein and DNA through certain signal transduction pathway. This Protein-protein interaction (PPI) and protein-DNA interaction (PDI) data helped us to port Human signal transduction pathway which is activated by FGF to fission yeast. Without knowledge of PPI and PDI through FGF signal pathway, we must undergo many trials and errors of adding and removing of proteins and promoters to form working signal transduction pathway. With the data from PID, we decided to port the FGF->FGFR1->STAT1->GAS pathway from human to fission yeast.
NCBINCBI provides biologists with many data. We used protein sequences and DNA sequences from NCBI nucleotide database and protein database. DNA sequence is very important to us because gene sequence of original protein is required to synthesize novel engineered protein. Even we don't synthesize gene, we should know the sequence of gene because BioBrick requires not only nucleotide material, but also its sequence information. NCBI also provided us with sequences of some antibodies to make single chain antibody sequence, and many Journals through PubMed service.
RCSB PDBRCSB PDB (Protein Data Bank) provides the data of structure of protein or other biomolecules. Key feature of data from PDB is structure. NCBI or Uniprot provide the sequence of proteins, but it does not show us their 3D-structures. With the structural similarity between FGF binding domain of FGFR and single chain antibody 16A1, we can ensure that the replacement of FGF binding domain with 16A1 to detect MPT51 is appropriate.
ToolsDuring our project, we processed much bio-information. It is not easy to process that massive information manually, so we used many bioinformatics tools for our projects. We marked restriction sites to select proper restriction enzyme, searched the nucleotide which codes query peptide sequence to find the coding region of certain genes, virtual-translated given nucleotide sequence to check if our sequence codes expected protein, and predicted and aligned structure of single chain antibodies with FGF binding domain of FGFR to check single chain antibodies are structurally similar to confirm that replacement of FGF binding domain with single chain antibodies is appropriate. We used BioEdit to mark restriction site, BLAST to find the coding region of certain genes and to compare similar proteins, Transeq for virtual-translation, Modeller for structural prediction of single chain antibody, and Matt for structural alignment between single chain antibodies and FGF binding domain of FGFR. BioEditBioEdit is the program for displaying biological sequences. It displays different amino acids or nucleotides in different colors to allow its users to check the change between sequences. It has many simple but useful functions. We used BioEdit for marking the restriction sites on given sequence to select proper restriction enzyme, which does not restrict coding region of gene. Other functions of BioEdit like phylogeny making or front-end of ClustalW are not used in our project, but they are also useful.
BLASTBLAST is the alignment search tools for protein or nucleotide sequences. There are five modes of BLAST; blastn (nucleotide to nucleotide), blastp (Protein to Protein), blastx (nucleotide to protein), tblastn (protein to nucleotide), and tblastx (translated nucleotide to translated nucleotide). We used tblastn to find the location of coding region of given protein and blastn to find the differences between transcription variants of same genes.
TranseqIt is not difficult to make protein sequence from nucleotide sequence. With codon table, we can make protein sequence manually without any special ability. To translate long nucleotide sequence, however, is not easy. Therefore, we used Transeq to virtually translate given nucleotides sequence. Transeq also do virtual-translation with shifted reading frame or non-transitional translation code tables like mitochondrial translation code table.
ModellerModeller is the program to predict the structure of protein with given peptide sequence based on the homology model. When given query sequence, Modeller searches for the similar sequence from database sequence whose structure is known. It assumes that similar sequences have similar structures and predicts the structure of query protein as the combination of known structure with similar sequence. This method is very useful for prediction of single chain antibodies because the structures of many original antibodies are known.
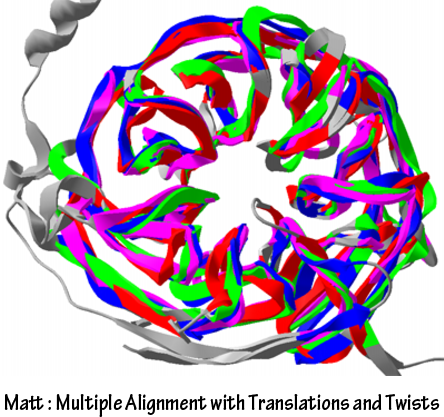
MattIt is also possible to compare the structures between FGF binding domain of FGFR and single chain antibodies by using Modeller manually, but it is not quantitative and estimated by the rule of thumb. So, the result is not useful for further analysis. We used Matt to compare the structures of FGF binding domain of FGFR and single chain antibodies. Matt uses algorithms to maximize shared structure with small translation and rotations. Matt provides quantitative results to enable estimation of similarity and aligned structures of proteins to visualize the alignment.
PyMolStructure of complex of protein and other biomolecules are often saved as format of “*.PDB”. Visualization and analysis of that structure and sequence are also important to design novel engineered proteins. PyMol is used for such purposes. We used PyMol for two processes. First of all, it is used to confirm iG-like regions of FGFR are really FGF binding domain. We downloaded the FGF binding domain of FGFR from RCSB PDB and checked the sequence binds to the FGF is really marked as iG-like regions. (Interleukin receptors have iG-like regions, but they do not bind to their signal molecules.) Other process using PyMol is to visualize the structural alignment result made by Matt. PyMol saves the image of biomolecules in png format.
Other Media Tools
References
|
 "
"