Team:TU Delft/16 July 2010 content
From 2010.igem.org
Contents |
Lab work
Ordered DNA + Solvent Tolerance and Hydrocarbon Sensing
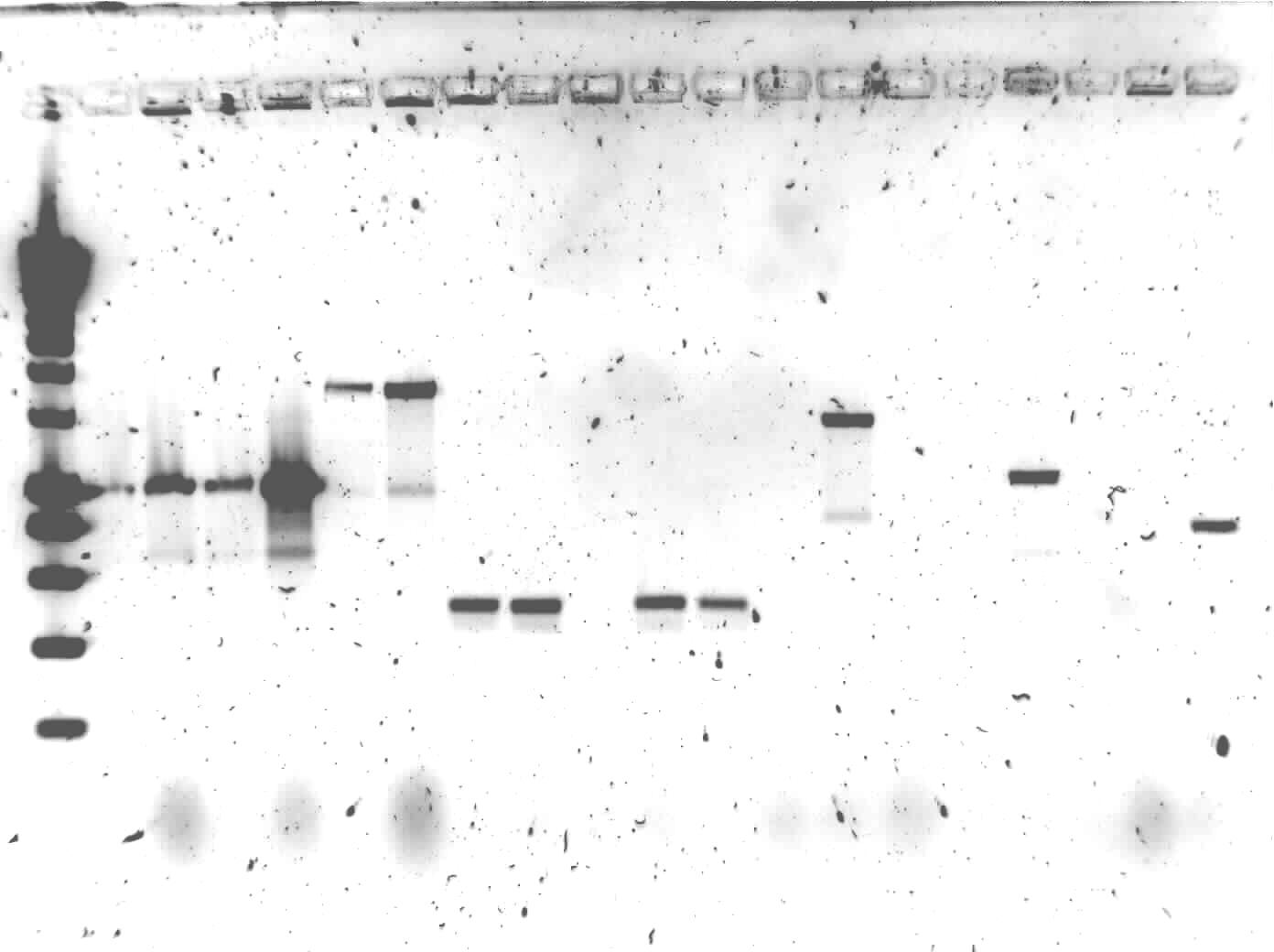
The colony PCR of yesterday was put on1% agarose gel
Lane description:
| # | Description | Expected lenght (bp) | Primers | Status |
| 0 | SmartLadder | n/a | n/a | n/a |
| 1 | transformant #1 of ligation mix AlkS + pSB1C3 | G00100 + G00101 | ✗ | |
| 2 | transformant #2 of ligation mix AlkS + pSB1C3 | G00100 + G00101 | ✗ | |
| 3 | transformant #3 of ligation mix AlkS + pSB1C3 | G00100 + G00101 | ✗ | |
| 4 | transformant #4 of ligation mix AlkS + pSB1C3 | G00100 + G00101 | ✗ | |
| 5 | transformant #1 of ligation mix ladA + pSB1C3 | G00100 + G00101 | ✗ | |
| 6 | transformant #2 of ligation mix ladA + pSB1C3 | G00100 + G00101 | ✗ | |
| 7 | transformant #1 of ligation mix RubA3 + pSB1C3 | G00100 + G00101 | ✗ | |
| 8 | transformant #2 of ligation mix RubA3 + pSB1C3 | G00100 + G00101 | ✗ | |
| 9 | transformant #3 of ligation mix RubA3 + pSB1C3 | G00100 + G00101 | ✗ | |
| 10 | transformant #4 of ligation mix RubA3 + pSB1C3 | G00100 + G00101 | ✗ | |
| 11 | transformant #5 of ligation mix RubA3 + pSB1C3 | G00100 + G00101 | ✗ | |
| 12 | transformant #1 of ligation mix PhPFDbeta + pSB1C3 | G00100 + G00101 | ✗ | |
| 13 | transformant #1 of ligation mix AlnA + pSB1C3 | G00100 + G00101 | ✗ | |
| 14 | transformant #2 of ligation mix AlnA + pSB1C3 | G00100 + G00101 | ✗ | |
| 15 | transformant #1 of ligation mix OprG + pSB1C3 | G00100 + G00101 | ✗ | |
| 16 | transformant #2 of ligation mix OprG + pSB1C3 | G00100 + G00101 | ✗ | |
| 17 | transformant #3 of ligation mix OprG + pSB1C3 | G00100 + G00101 | ✗ | |
| 18 | transformant #1 of ligation mix PhPFDalpha + pSB1C3 | G00100 + G00101 | ✗ | |
| 19 | transformant #2 of ligation mix PhPFDalpha + pSB1C3 | G00100 + G00101 | ✗ |
We harvested the 1 mL bacterial cells of possible correct transformations. The pellets were stored at -20 °C. We used 3 mL of the bacterial cells to make -80 °C stocks.
Characterization of Anderson RBS sequences
Fluorescence measurements Attempt #2
Data analysis to follow shortly (good stuff!)
Assembly of reference construct & positive control
Yesterday's digestion products of K081005 and I13401 were set for ligation over the weekend at 4 °C:
| # | BioBrick | Fragment | Recipient plasmid | Final volume |
| 1 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K136011 K136011] | 17.5 μL ‘E-K081005-S’ | 4 μL ‘E-pSB1A2 I13401-X’ | 25 μL |
| 2 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K136011 K136011] | 17.5 μL ‘E-K081005-S’ | 4 μL ‘E-pSB1A2 I13401-X | 25 μL |
| 3 | Ligation control | None | 4 μL ‘E-pSB1A2 I13401-X | 25 μL |
To all samples with an end volume of 25 μL, 2.5 μL Ligase buffer was added.
The Birnboim method was used to isolate J23100 and I13522 (both in pSB1A2) from the o/n 5 mL LB cultures. We used 3 mL of the bacterial cells to make -80 °C stocks . The following concentrations of plasmid were obtained:
| BioBrick | Concentration (ng/μL) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_J23100 J23100] | 130.7 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 I13522] | 106.7 |
 "
"