Team:SDU-Denmark/labnotes3
From 2010.igem.org
Group: Flagella
Incertion of Promoter + RBS in pSB3T5
Done by: Christian and Louise
Parts used: J13002 (promoter+RBS) and pSB3T5
Restriction and Gel extraction
Protocol: [RD1.1]
Notes: We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. We also added 10 ul less water and 10 ul more PCR product than discribed in the protocol
Restriction mixture
- 38 ul Water
- 4 ul EcoRI enzyme
- 4 ul PstI enzyme
- 8 ul Fast Digest green buffer
- 30 ul PCR product (Freeze tube white 25 PROMOTER + RBS)
OR
- 30 ul PCR product (Freeze tube white 29 PLASMID)
Loading:
2 x 21 ul J13002 was loaded on a 2% gel with a 100-1000bp ladder. The Restricted J13002 is 95bp.
2 x 21 ul pSB3T5 was loaded on a 1.5% gel with a 100-10,000 ladder. The restristricted plasmid is 3215bp.
Result:
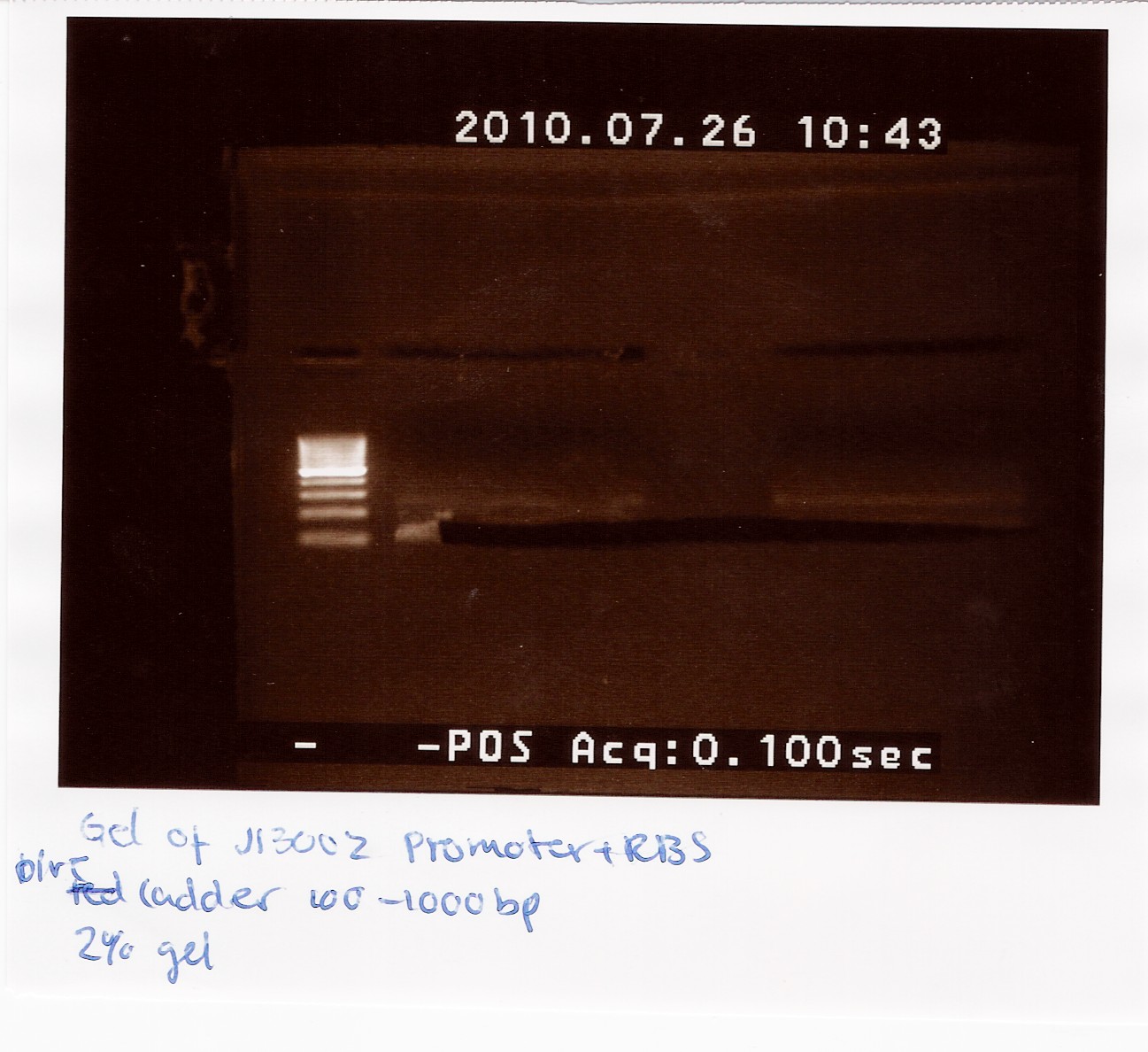
The picture shows a band around 100bp.
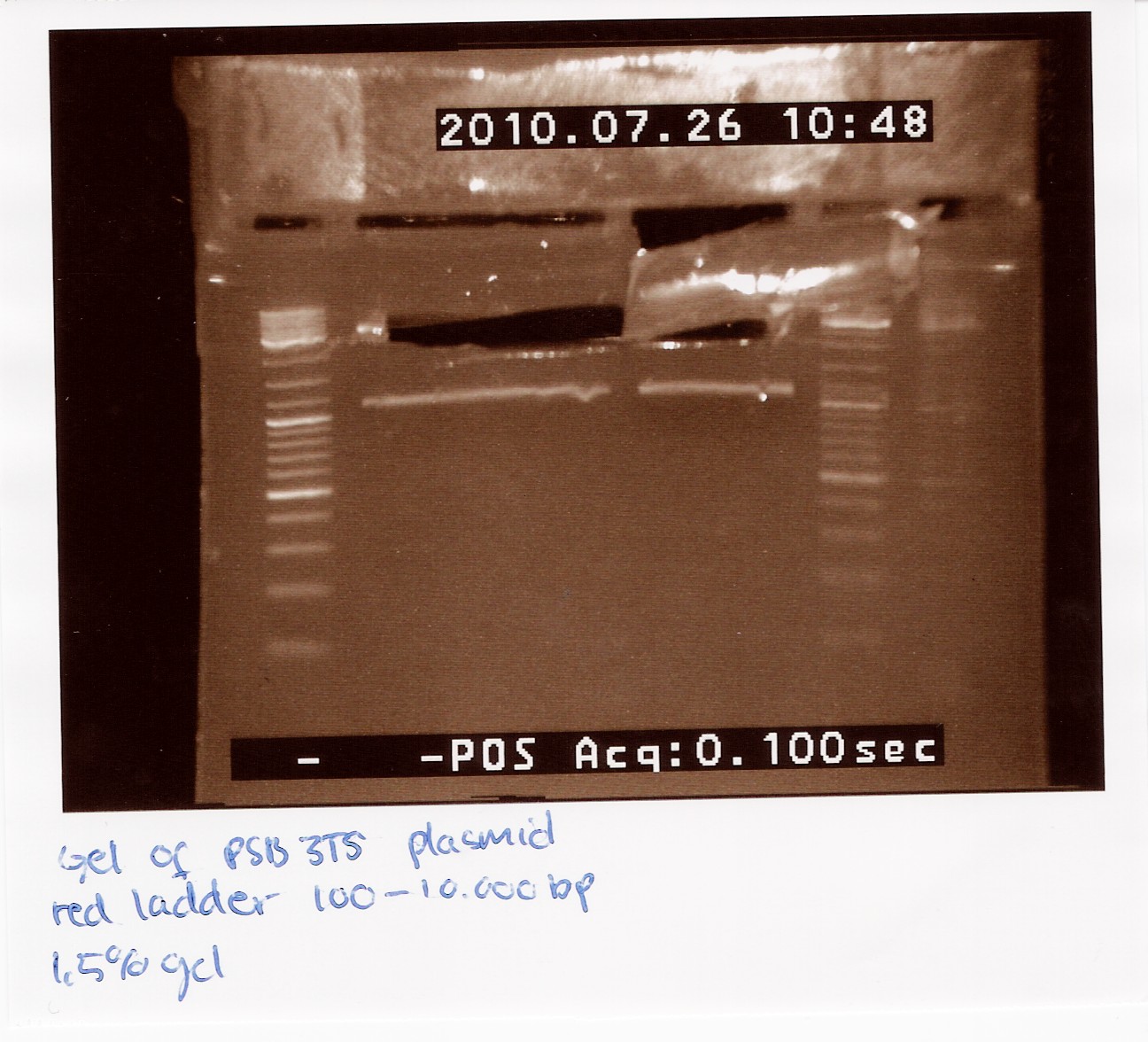
The picture showes two bands, one around 3000bp which is the plasmid and one around 1000bp, which is RFP.
Purification of J13002 and pSB3T5
Protocol:
Notes: four tubes were marked and weighed (all had a mass of 1g). Bands were cut from the restriction gels, added to the tubes and weighed.
| Tubes |
1P (Plasmid) |
2P |
3B (Brick) |
4B |
| Tube weight |
1g |
1g |
1g |
1g |
| Gel weight |
170mg |
110mg |
245mg |
160mg |
Sample capture: 300 ul Capture buffer type 3 was added to the tubes and the tubes were placed at 60 degrees for 20 minutes.
Sample binding:The Capture buffer sample mix were added to MicroSpin Columns, stood for 1 minute and centrifuged for 30 sec. at 16,000g.
The Wash and Dry was carried out according to the protocol.
Elution: 10 ul Elution buffer type 6 was added to the columns, stood for 1 minute and centrifuged for 1 minute at 16,000g. The columns were thrown out and NanoDroped.
NanoDrop:
| Tube |
Concentration (ng/ul) |
| 1P |
9.6 |
| 2P |
2.9 |
| 3B |
6.7 |
| 4B |
3.2 |
The Concentrations were quite low, but we pooled them (1P+2P and 3B+4B) and used them for ligation.
Pfu PCR of J13002 and B0015
Done by: Louise
Date: July 28th
Protocol: [CP1.1]
Notes:
5 x J13002 and 5 x B0015 were made.
Pre-mix x 10
400 ul water
50 ul Pfu-buffer + MgSO4
15 ul dNTP
15 ul VF2 primer
15 ul VR primer
5 ul Pfu polymerase
Total volume: 500 ul
To each PCR-tube 50 ul pre-mix and 2 ul DNA was mixed and the tubes were loaded on the PCR maschine.
PCR program
| Progress |
Temp. (c elcius) |
Time (min.) |
| Start |
94 |
3 |
| Denaturing |
94 |
2 |
| Annealing |
55 |
0.5 |
| Elongation |
72 |
0.5 |
| GOTO |
rep x 29 |
|
| End |
72 |
2 |
| Hold |
4 |
--Louch07 09:38, 28 July 2010 (UTC)
Pfu PCR of FlhDC with mutation
Done by: Louise
Date: July 28th
Protocol: [CHP1.1]
Notes: Template sample: Freeze tube white 31.
Six samples were made.
Pre-mix x 6
240 ul water
30 ul Pfu-buffer + MgSO4
9 ul 10mM dNTP mix
9 ul FlhD fw primer
9 ul FlhC rev primer
3 ul Pfu polymerase
Total volume: 300 ul
In the PCR tubes 50 ul pre-mix and 2 ul template was mixed and loaded into the PCR machine.
PCR Program:
| Progress |
Temp. (c elcius) |
Time (min.) |
| Start |
94 |
3 |
| Denaturing |
94 |
2 |
| Annealing |
55 |
0,5 |
| Elongation |
72 |
1 |
| GOTO |
rep x 29 |
|
| End |
72 |
2 |
| Hold |
4 |
--Louch07 09:28, 28 July 2010 (UTC)
Follow-up colony PCR
Date: 26/7
Methods: Colony PCR
Protocol: CP1.3
Experiment done by: Maria, LC
Notes: We made a sample for every plate from the last colony PCR. Out of the 29 samples we could only run 25 at once in the PCR machine. Plate 23 was missing, so there is no sample for it.
Results: 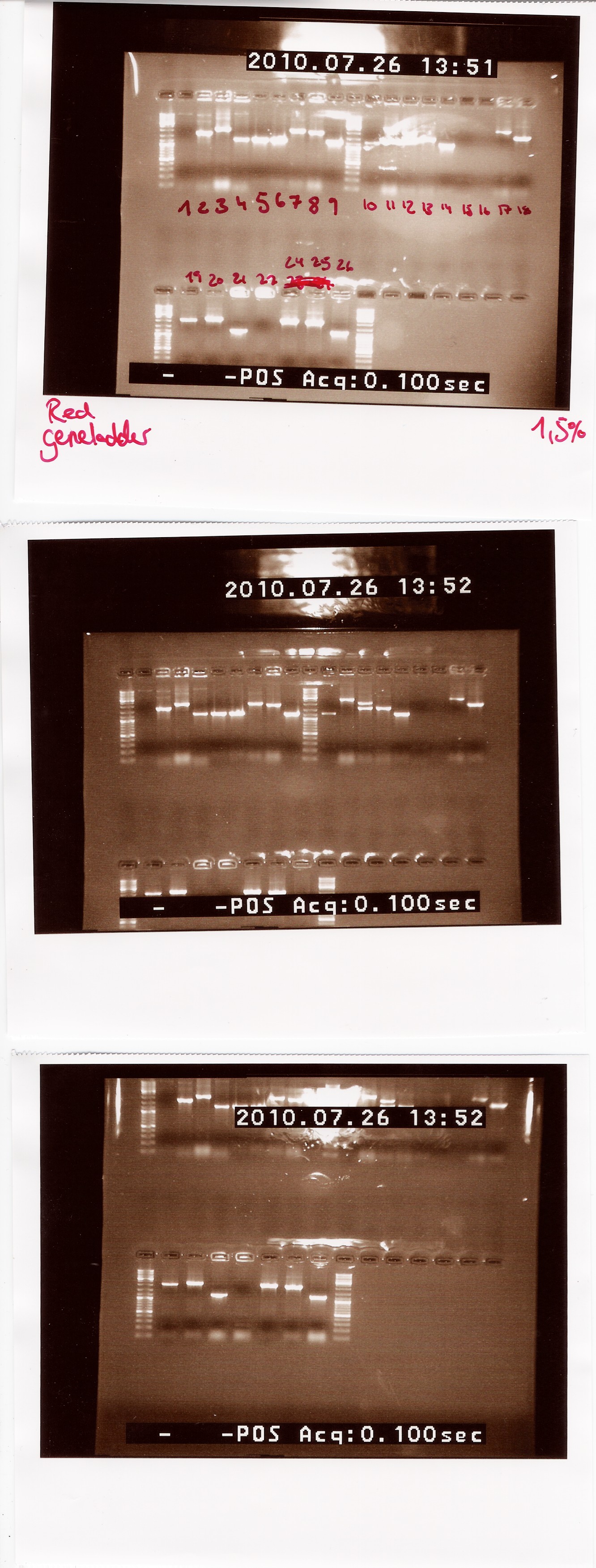
Lengths of PCR products:
1200: 4, 5, 6, 9, 10, 14, 21, 26
1500: 13
1750: 2
1900: 8, 25
2000: 3, 7, 12, 18, 19, 24
2200: 20
2500: 11, 17
Colonies 1, 15, 16 and 22 gave no result.
Analysis: Since we had so many different lengths, we will cut one from each length, specifically colony: 2, 8, 11, 13, 20, 24, 26.
Restriction digest of PCR product from 26/7
Date: 26/7
Methods: Restriction digest
Protocol: RD1.1
Experiment done by: Maria, LC
Notes: We took one of each length PCR product and cut them with XbaI and PstI. The especially interesting one was sample 26, since we suspected it to be a succesful ligation. If it is FlhD,C we would expect it to get cut like this:
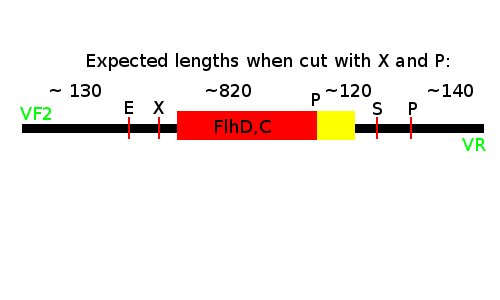
We loaded the cut and uncut sample next to each other. They got loaded as follows and RFP was used as a control:
| Lane |
Sample |
Cut/Uncut |
| 1 |
2 |
c |
| 2 |
2 |
u |
| 3 |
8 |
c |
| 4 |
8 |
u |
| 5 |
11 |
c |
| 6 |
11 |
u |
| 7 |
13 |
c |
| 8 |
20 (loading error) |
u |
| 9 |
20 |
c |
| 10 |
13 (loading error) |
u |
| 11 |
24 |
c |
| 12 |
24 |
u |
| 13 |
26 |
c |
| 14 |
26 |
u |
| 15 |
7(RFP) |
c |
| 16 |
7(RFP) |
u |
Results: 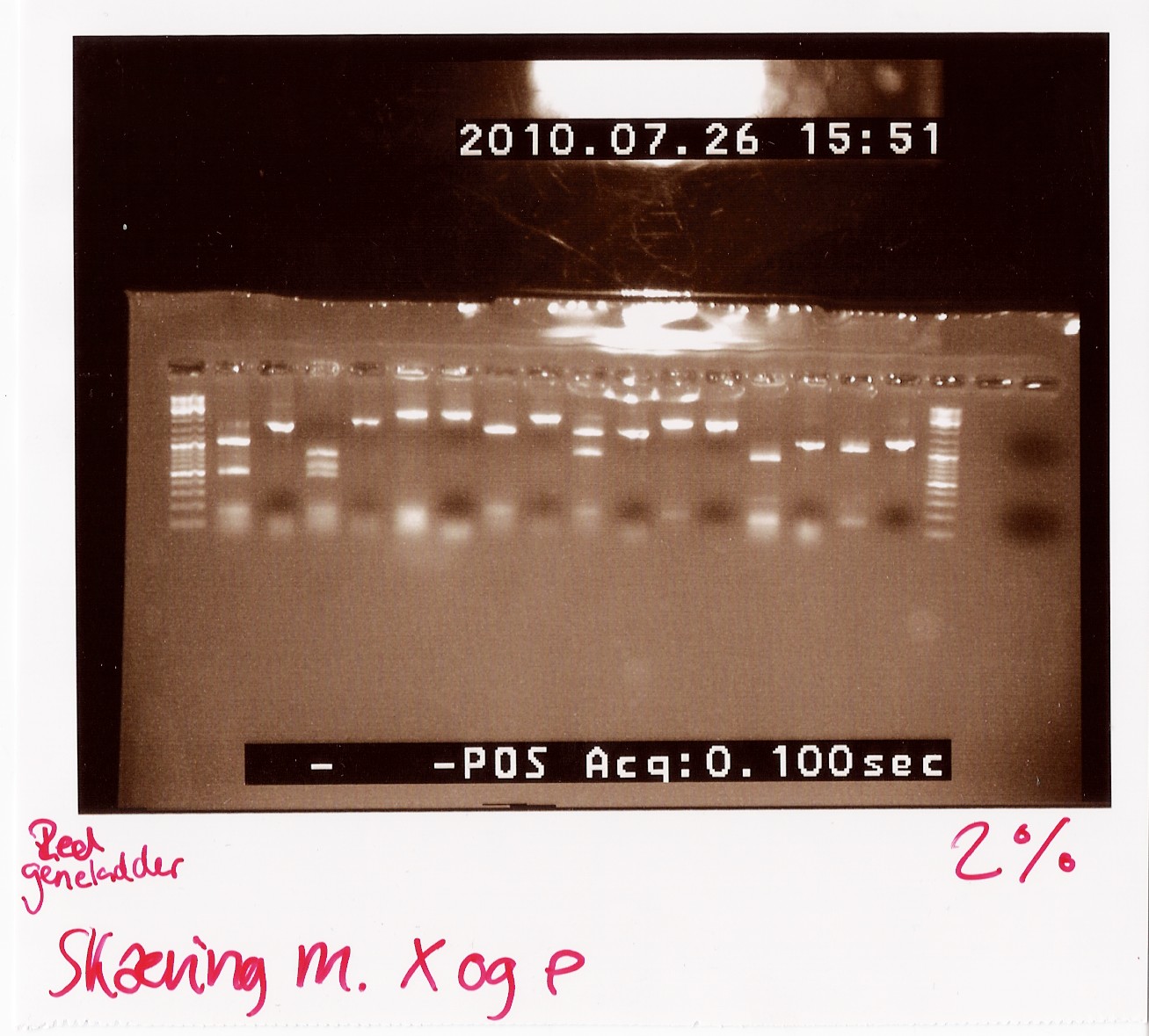
Analysis: Sample 26 looks very close to the right lengths, so we will prepare some of the 1200 BP length PCR products for sequencing.
Miniprep of follow-up colony PCR
Date: 27/7
Methods: ON, Miniprep
Protocol: MP1.1
Experiment done by: LC
Notes: Samples L4, L5, L6, L9, L10, L14, L21 and L26 (L for ligation from the follow-up colony PCR) miniprepped and NinaB samples from the 4 frozen cultures of it.
Results: 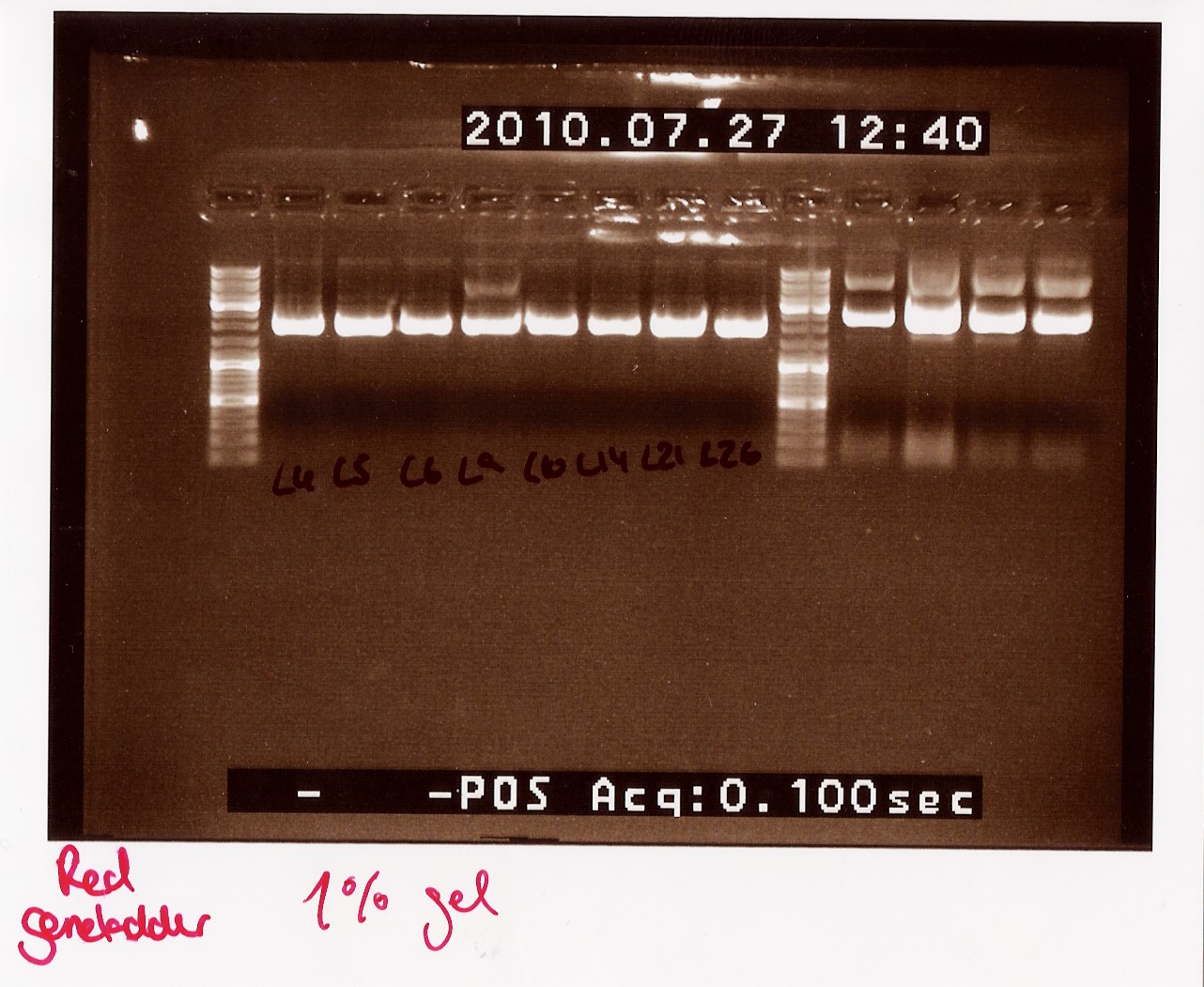
All ligations were around the same length, just under 2000 BP. Even though the correct length should have been about 1000 BP longer, this is not surprising since uncut plasmids often show 1000 BP less in length. NinaB was also too short and there was a second band showing higher up in the gel (weird!). The concentrations averaged around 80ng/ul for the samples.
Analysis: We decided to do a new miniprep where we start with a higher concentrated overnight culture, which we will achieve through boosting the ON before doing the miniprep.
Restriction digest of PCR product from 26/7
start date: 27/7
Methods: Restriction digest, gel electrophoresis
Protocol:RD1.1
Experiment done by: Maria, LC
Notes:
All colonies with PCR products of 1200bp (see Follow up coloni PCR) was digested with XbaI and PstI.
Coloni #4,5,6,9,10,14 and 21 was selected.
Digested RFP PCR product (#6 from Coloni PCR) was used as controle.
If it is FlhD,C we would expect it to get cut like this:

Digested samples were loaded along with undigested ones. Samples were loaded onto a 2% gel. Hyper ladder 2 was used as marker.
Loading scheme:
| Lane |
Sample |
Cut/Uncut |
| 1 |
4 |
c |
| 2 |
4 |
u |
| 3 |
5 |
c |
| 4 |
5 |
u |
| 5 |
6 |
c |
| 6 |
6 |
u |
| 7 |
9 |
c |
| 8 |
9 |
u |
| 9 |
10 |
c |
| 10 |
10 (loading error) |
u |
| 11 |
14 |
c |
| 12 |
14 |
u |
| 13 |
21 |
c |
| 14 |
21 |
u |
| 15 |
6(RFP) |
c |
| 16 |
6(RFP) |
u |
Results:
gel electrophoresis:
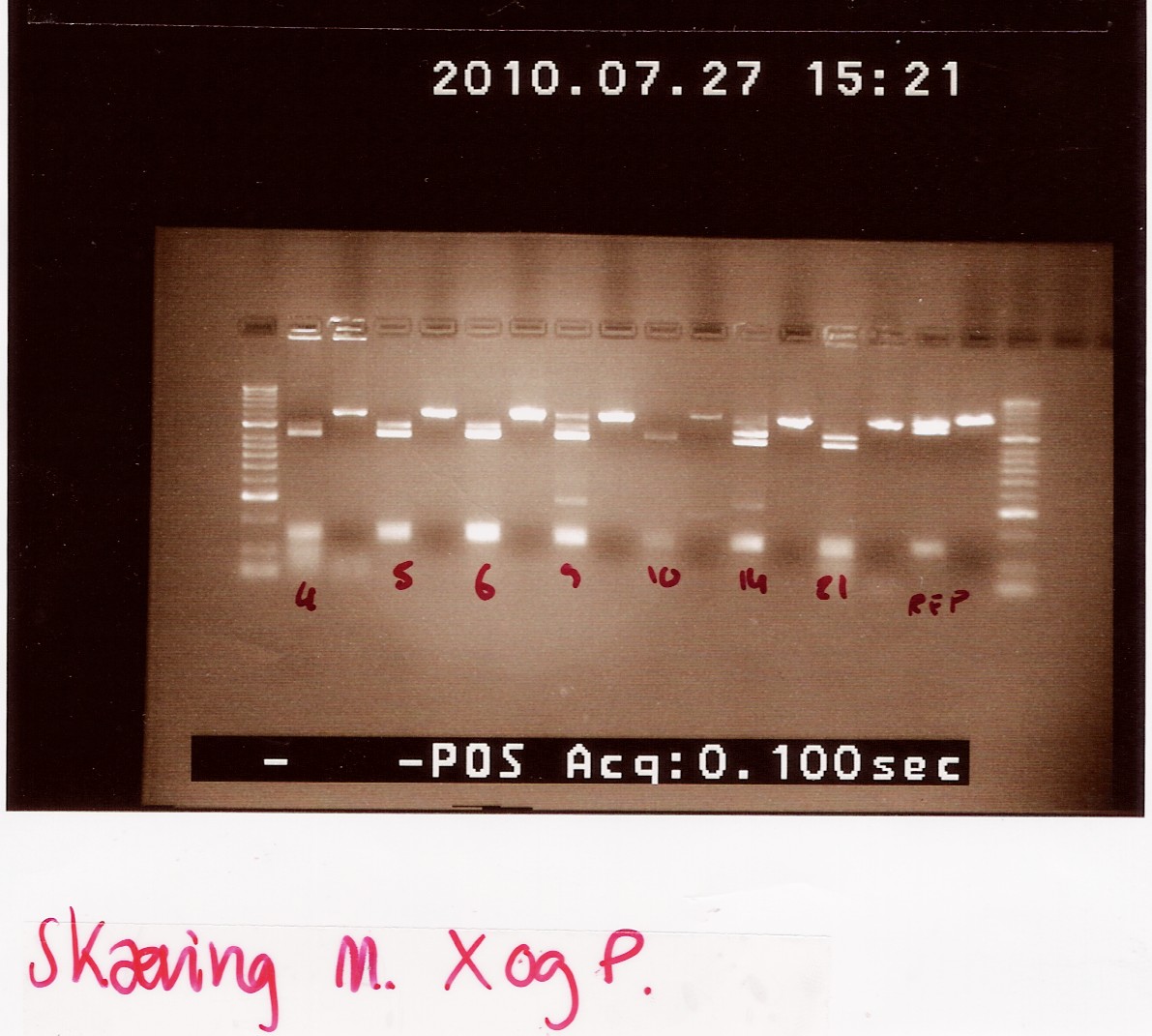
Analysis:
The lanes containing digested PCR product from coloni #5, 6 and 21 have a band smear around 100-200bp,and a band at around 800bp.
This may indicate correct inserted flhD/C.
ON cultures are made of each of these colonies to use for boost miniprep.
--Tipi 13:26, 28 July 2010 (UTC)
Digestion and gel extraction of pSB3C5 and J13002
Done by: Pernille
Date: the 26th of july
Protocol: [RD1.1] and gelpurificaton (DE1.3)
Notes:because the concentration for both plasmid and biobrick is rather small I dobbelt the volumen of the reagent compared to the protocol. in the soultion contaning the brick the volumen of the PCR product was triple and to adjust the total volumen i added 5ul less water. We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. both the plasmid and brick was cut at the E and P site. After the cutting the plasmid and brick was run on seperate gels because of there different size. the plasmid on a 1,5% gel and brick on a 2% agarose gel. When I observed the gel under the UV lamp the was no visible band on the gel were the brick was loaded. therefore it was only possible to do DNA extrated from the gel contraining the plasmid. The Dan was eluted in 10ul of water and I eluted two times. The DNA concentration was measured on the NaonDrop and was found to 55,41ng/ul for the first elution and 28,03ng/ul for the second. the to samples are pooled and stored in a frezze tube.
 "
"