Team:Brown/Project/Ecargo/Experimental Design
From 2010.igem.org
E.Cargo
Experimental Design
CONSTRUCTING TAT-TF
- The Tat-PTD sequence was obtained by our lab as part of a potential cell labelling project that was later dropped in the summer. We initially ordered the Tat domain with an scFv (single chain variable fragment antibody) conjugated to nuclear histone proteins. Our intention was to express this Tat-scFv fusion, purify, label with rhodamine, and apply to mammalian cells. We would've expected fluorscence to localize to nuclear histones, confirming that Tat was carrying the antibody through a live cell membrane.
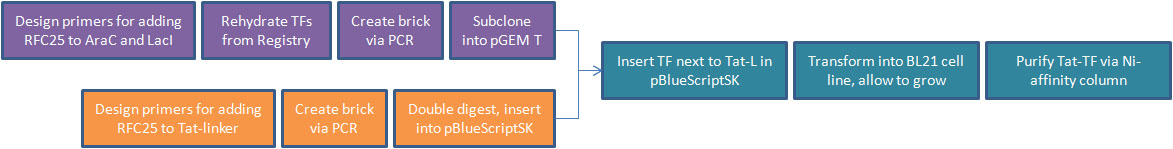
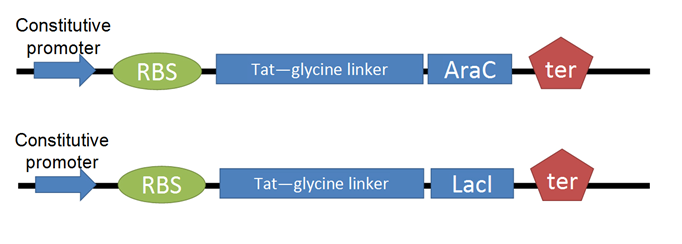
- After shifting directions in our research, we decided to pursue Tat-PTD applications involving transcription factors. For our Tat-TF, we needed to create the Tat-PTD with RFC25 ends (the Freiburg assembly standard, optimized for protein fusion). We used PCR to attach sites to the existing circular Tat DNA, avoiding the process of resynthesizing. This was biobricked as our Tat-linker.
- The next step was to adapt the transcription factor for use with Tat. We decided to use two TFs from the registry, [http://partsregistry.org/Part:BBa_I732100 LacI] and [http://partsregistry.org/Part:BBa_C0080 AraC]. We then made a series of modifications to our proteins through PCR.
- We removed an existing LVA tag, a 3 a.a. sequence that increases rate of proteolytic degradation, because our TFs were meant to be isolated and have a long lifespan. We affixed a 6-histidine tag to facilitate purification with Nickel-affinity columns. This was done on the C-terminus to avoid isolation of incompletely translated peptides. Finally, we converted our BioBricks to RFC25.
- The linear PCR products of the transcription factors were then subcloned into pGEM-T Easy. The PCR product of the Tat-linker was double-digested and inserted into pBlueScriptSK.
- We cut the transcription factor out of pGEM-T Easy, run out on a gel to confirm successful cut, extract bands, then insert alongside the Tat-linker in pBlueScriptSK using sites for protein fusion in RFC25.
- We inserted Tat-TF/pBlueScriptSK into competent E. coli (BL21 strain) and use ampicillin resistance to select for successful transformants. NOTE: This represents the farthest we have reached in this section of our experiment.
- We will grow an overnight liquid culture of successful transformants, harvest cells and use Ni-affinity column to isolate purified Tat-TF protein.
TESTING THE TAT-TF
- To test for efficacy of Tat-TF control, we decided to use two reporter constructs. The [http://partsregistry.org/Part:BBa_J04421 LacI reporter construct] was taken straight from the registry. The AraC reporter construct was assembled from registry parts:
1. Production of CyanFP is controlled by pLacI, which is constitutively on and repressible by addition of LacI protein.
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0020 Cyan Fluorescent Protein]
- [http://partsregistry.org/Part:BBa_K091110 LacI repressible promoter]
2. CherryFP production is linked to pAraC, which is inducible by addition of the AraC protein.
- [http://partsregistry.org/Part:BBa_R0080 AraC inducible promoter]
- [http://partsregistry.org/wiki/index.php/Part:BBa_J06702 RBS-Cherry Fluorescent Protein-terter]
- We then transformed each reporter construct into E.Coli (XL1B) and froze them into glycerol stocks for future use.
 "
"