Team:Berkeley/Clotho
From 2010.igem.org
Clotho
- Home
- Project
- Parts
- Self-Lysis
- Vesicle-Buster
- Payload
- [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Berkeley Parts Submitted]
- Results
- Judging
- Clotho
- Human Practices
- Team Resources
- Who We Are
- Notebooks:
- [http://www.openwetware.org/wiki/Berk2010-Daniela Daniela's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Christoph Christoph's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Amy Amy's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Tahoura Tahoura's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Conor Conor's Notebook]
The UC Berkeley iGEM Team of 2008 started the Clotho software project to provide a suite of software tools for synthetic biology. Since then, Clotho has evolved into a platform on which new software tools can be developed. Clotho as a platform factors out the common functionality that many software tools would provide, thus allowing new tools to be developed more quickly and making it easy to integrate new tools with existing tools.
This summer, the 2010 Berkeley iGEM Team is proud to release Clotho 2.0, featuring the robust Netbeans plugin platform, a reworked framework for automating workflows, as well as a shift from dynamic data models to a static Clotho data model. Read more about these new features here on our wiki, or visit our new public [http://www.clothocad.org/ website] for more information and to download our wonderful tool!
Plugin Platform
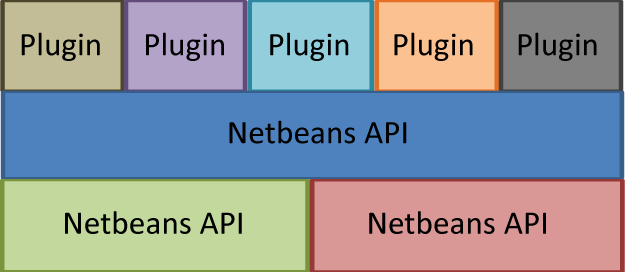
Clotho provides a plugin framework for the development of new tools. As illustrated by the picture below, Clotho plugins are built on top of the Clotho API, and are able to use functionality provided by this API. Clotho in turn uses functionality from other libraries below it. The plugin framework is built on the Netbeans Platform ([http://netbeans.org netbeans.org]), which manages the plugin environment. This includes checking that all the dependencies of a plugin are satisfied before it is launched, managing all the files that may be necessary for a plugin to function correctly, as well as actually loading the executable code for each plugin into Clotho at runtime. The Hibernate library provides a structured interface to external databases and is discussed in more detail on the Data Model page.
Clotho classifies plugins into eight distinct categories: ClothoAlgorithm, ClothoConnection, ClothoFormat, ClothoGrammar, ClothoPlugin, ClothoTool, ClothoViewer, and ClothoWidget. These categories are based on the different types of functionality a plugin can provide. For example, a ClothoConnection provides connections to a database, while a ClothoTool is a tool built on the Clotho platform. Plugins are discovered and loaded dynamically when Clotho starts. For more information about how to write a plugin, what functionality the Clotho API provides, and to download plugins for your own use, visit the [http://clothocad.org clothocad.org].
Automated Workflows
Automating synthetic biology workflows has always been a goal of the Clotho project. As devices become increasingly complex and utilize greater numbers of parts, it makes more and more sense to automate many of the steps in the construction of these devices. Last summer, the Berkeley iGEM team sought to build an automation framework for synthetic biology on top of Kepler, a tool for automating scientific workflows developed by the CHESS group at Berkeley ([http://chess.eecs.berkeley.edu/ chess.eecs.berkeley.edu]). Kepler is a powerful tool, but its large size and complexity makes it hard to integrate with Clotho. Our attempts to do so were only mildly successful, so we decided to recode a simpler automation framework using the ideas from the Kepler project.
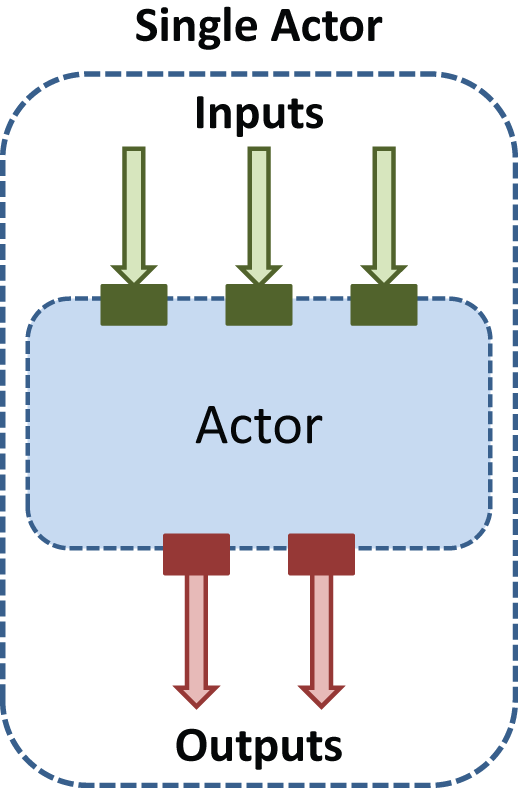
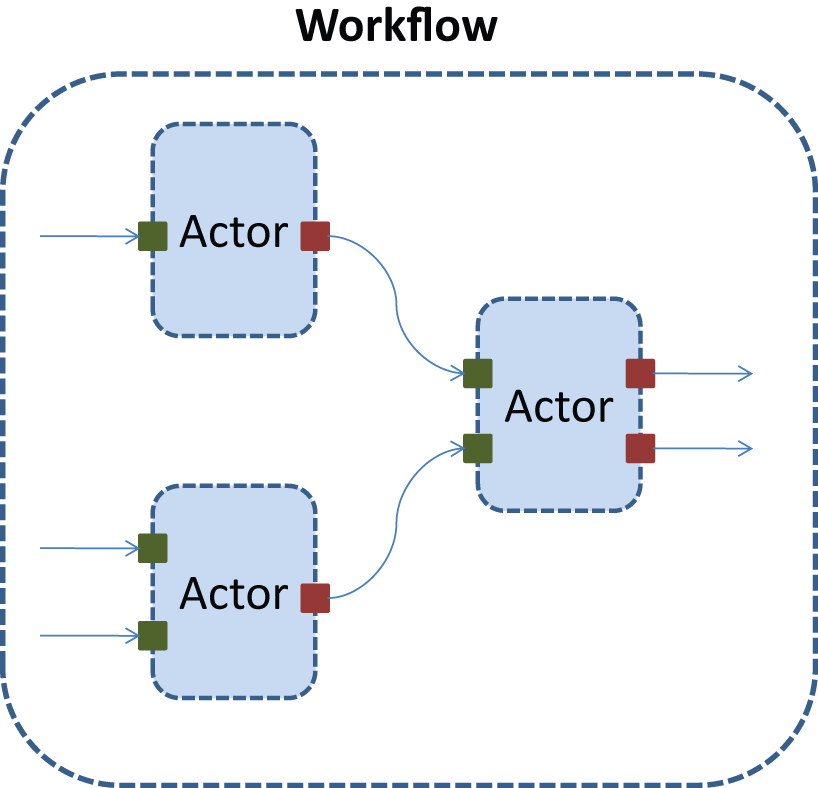
What we came up with is illustrated above. A unit of automation is called an Actor, which executes some algorithm. Each Actor defines a set of InputPorts and OutputPorts, and each Input/Output port only works with one type of data. Thus, the set of ports of an Actor specifies how it can interact with other Actors. Actors allow developers to reuse code from other developers, as well as allowing users to chain together Actors written by different developers into new workflows, customized for their own needs.
A workflow is built by connecting the inputs and outputs of multiple Actors. After a workflow has been constructed and fed initial data, Clotho will execute the Actors in the workflow according to the partial ordering defined by the workflow(which is a directed acyclic graph). Users can construct workflows using the scripting interface provided by Clotho. In addition, a graphical interface to build workflows is currently under development by the Clotho team. These tools will make it easy for users to share their automated workflows with the community.
Data Model
Potentially the most fundamental change this summer was a shift from dynamic data models to a static Clotho data model. In previous years, Clotho tried to remain data model agnostic by allowing users to define their own data model. However, this introduces major problems for developers trying to build tools for Clotho. If one tool is built assuming one data model, then it is unlikely to be compatible with another tool built assuming a different data model. Even worse, if a user changes the data model after installing a tool, the tool could be rendered nonfunctional.
Because we could see no good solution to the problems generated by a dynamic data model, the Berkeley iGEM decided to fix the data model that Clotho uses. Although this data model is still under development, tools can be guaranteed that the data model for one particular version of Clotho will not change, making it possible for tools to share data with each other. A static data model also allows Clotho to validate data before it is saved to a database and shared with other users, which is especially important in the light of biosafety. More information is available on our Human Practices page about how Clotho makes doing synthetic biology safer.
 "
"