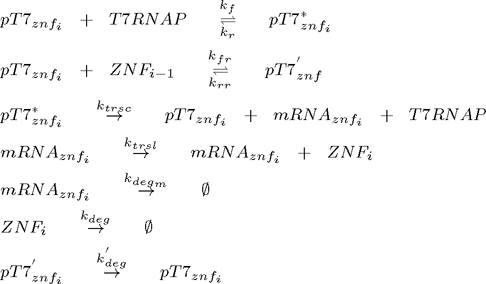Team:Slovenia/PROJECT/oscillator/repres
From 2010.igem.org
Contents |
Repressilator
Repressilator is an oscillator initially based on at three repressors connected into a cyclic feedback loop. The only experimentally tested repressilator contains three bacterial repressors: LacI, TetR and λ cI (Elowitz et al., 2000). In order to observe the oscillations of the repressilator a reporter gene that helps keeping track of the process is also included. Our selection of zinc fingers as artificial repressors provides the building blocks to construct new repressilators, including an extended number of genes in the cycle. Each zinc finger protein acts as repressor to the next gene in a sequence, forming a circular repression scheme presented in the Figure 1. Using the ZNF-based repressor platform we are no longer limited by the availability of natural repressors and having to match their specific properties, as the binding affinities of zinc fingers and their stability/lifetime can be very similar.
The figure presents the situation where three zinc fingers are used in a similar topology as the original repressilator. However, any odd number of zinc finger repressors can be used as long as we can keep the system alive (not over-saturating the cell). One of the zinc fingers also acts as a repressor of the reporter gene. All promoters were activated by the T7 RNA polymerase, allowing for the orthogonality of the system for prokaryotic and eukaryotic cells. Each of the promoters is however modified by the addition of binding site for the preceeding zinc finger in the cycle.
Because we can select very similar zinc fingers (in terms of the binding site), the same kinetic rates and constants were used for all of them. The following zinc finger initiation and dissociation rate values were used: k_on = 0.031 uM^-1 sec^-1, k_off = 0.0002 sec^-1 (Yang et al, 1995). Velocity of RNA polymerase used was 200 nucleotides/second (McAllister et al., 1997), while the T7 RNA polymerase initiation rate and dissociation rates were set to 50uM^-1 sec^-1 and 0.2 sec^-1 respectively (Kuzmine et al., 2002).
Since chemical equations are the same for all zinc fingers, the system can be represented as a set of following equations (pT7* is activated promoter, while pT7' is repressed promoter):
where i =1,2,3,...n and n is the number of zinc fingers. Additional equations for flourescent reporter protein under the control of T7 promoter and for T7 RNA polymerase under the control of constitutive promoter for T7 RNAP must be added:
Michael B. Elowitz, Stanislas Leibler, Departments of Molecular Biology and Physics, Princeton University, USA, 2000. A synthetic oscillatory network of transcriptional regulators.</p> Nature Vol 403: 335-338
Wei-Ping Yang, Herren Wu, Carlos F Barbas III, 1995. Surface plasmon resonance based kinetic studies of zinc finger-DNA interactions. Journal of Immunological Methods, 183: 175-182
McAllister, W. T., 1997. Transcription by T7 RNA polymerase. Nucleic Acids Mol. Biol. 11: 15–25
Iaroslav Kuzmine, Craig T Martin, 2002. Pre-steady-state kinetics of initiation of transcription by T7 RNA polymerase: a new kinetic model. J. Mol. Biol. 305: 559-566
 "
"