Team:USTC Software/User Interface
From 2010.igem.org
Contents |
Welcome to the world of creation and imagination!
|
As the world enters upon an age of human nature destructions, of resource starvation and the ignorance of ecological balance, the earth is more and more under the control of human beings, there is less room for the survival of animals, various kinds of microorganism are being killed and regenerated circulatory in laboratory. However, there exsits a special kind of bacterial, who is too diligent to be willing to be controlled by human, they use their magic power to learn certain skills to being survived successfully. Reviewing their developmental history, their ancestors made great contribution to the world under successive human experiments. They show us the meaning of Biobrick, they grasp the ability to adapt themselves better to the outside world, they know how to realize their value furthest and greatly, they are the E.COLI! Now let us begin the E.COLI magical mystery tour~~ |
Input
|
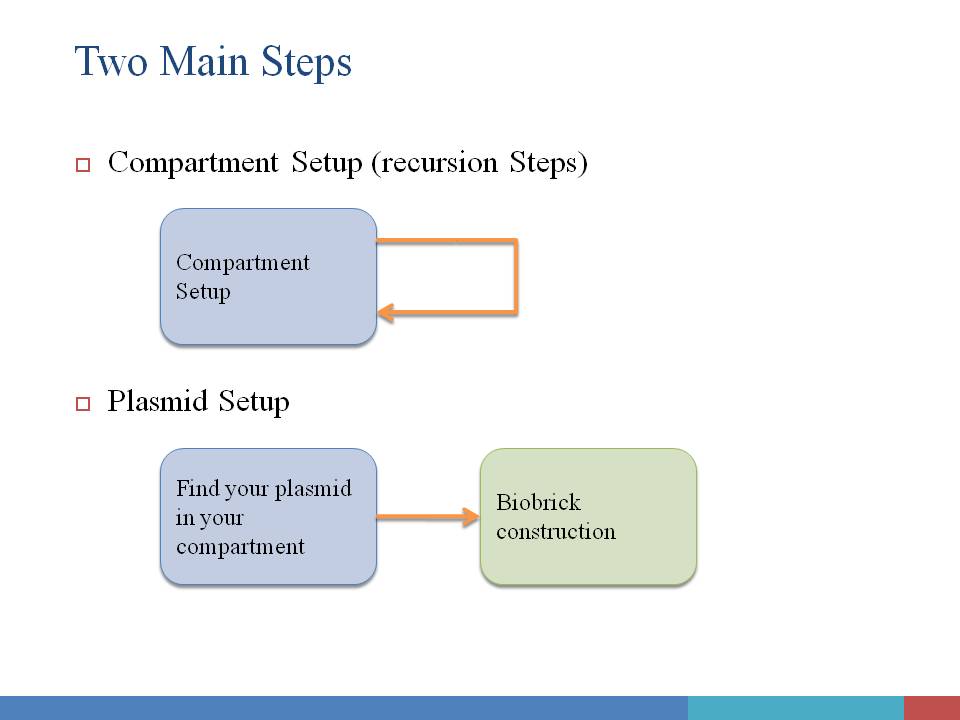
In order to give users a clear and definite goal on input. The whole input process is divided into two main steps: Compartment Construction and Plasmid Construction. In the first step, users only need to focus on the input of environmental condition, compartment, spatial structure of compartments, and the species contained in each compartment. In the second step, only plasmids are editable, and users can click on each of them to assemble biobricks into functional device. Of course, useful tips will be given along with your input process. We here give a design of our user interface of input. It is under construction. |
Step#1 Compartment Construction
|
| |
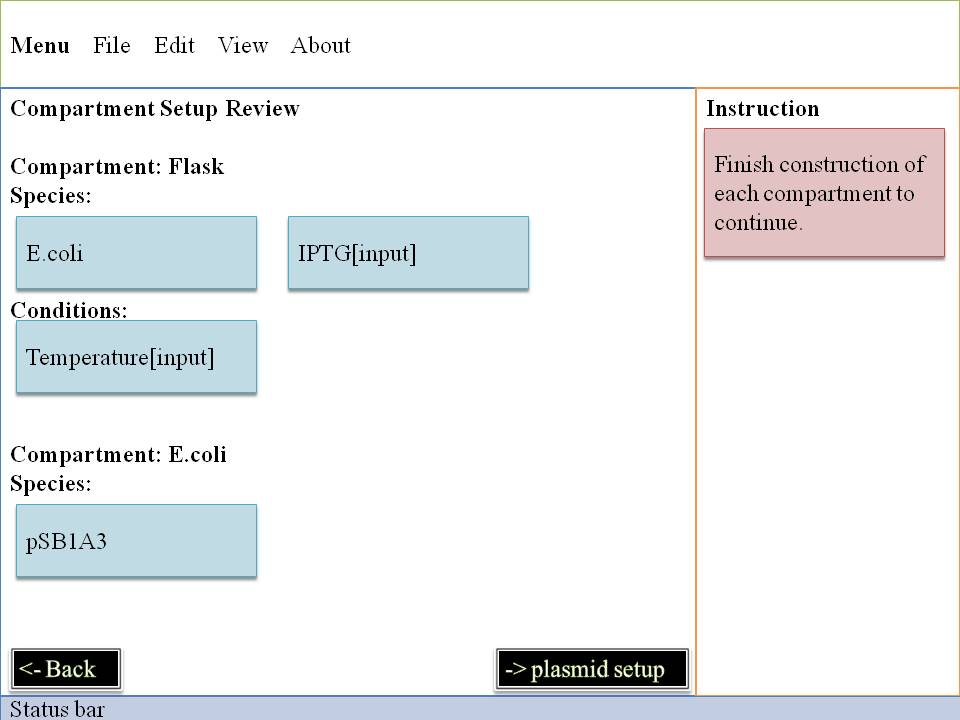
| Left:
When you enter in our interface,the first step is to choose the environment of our system . A flask is like a container , all the reactions are happening in it . After that you may pick a appropriate condition for our lovely E.Coli . One E.Coli isn't enough for some specific problem , so you have to decide how many you will pick up . Small molecule is also available in our interface , you can decide their amount per second . They can be static or changing all the time . It's up to you ! | Right:
You can choose E.Coli and set their parameters one by one . Plasmid must be set up in this level . So you can smoothly enter into next level to make you own code of genes~~ |
|
OK!Now it's the time to give our system some more data , write down your favourite condition , it's good to be god , huh?~ Take it easy , next part is more attractive ! |
|
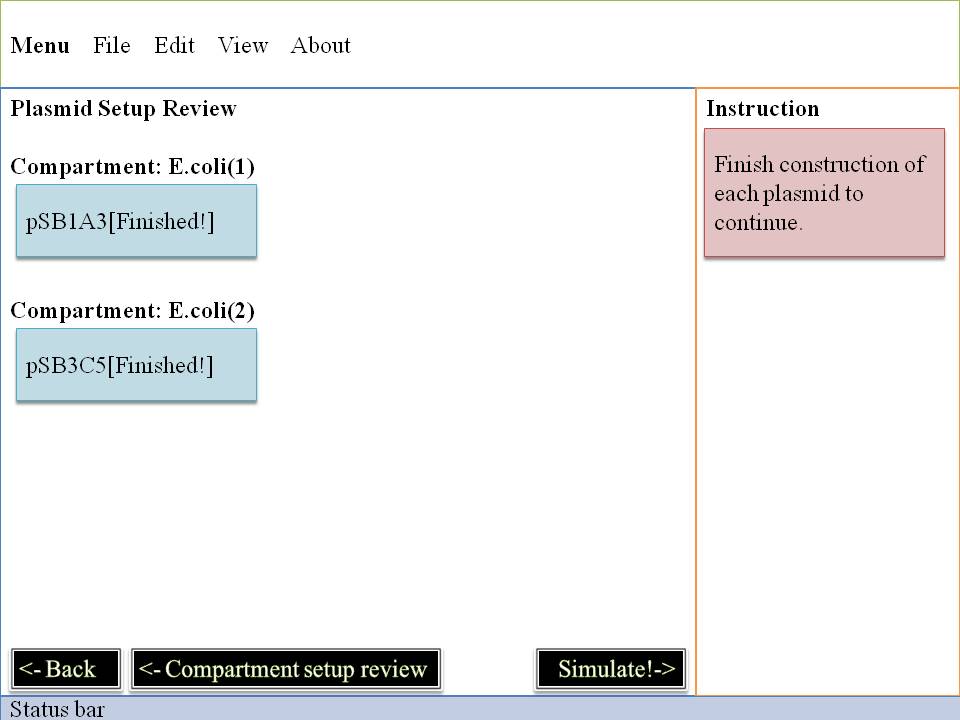
Step#2 Plasmid Construction
|
Here's the most interesting part! You can change the fundanmental construction of our cute Mr.ecoli! First you must locate on which E.coli you are interested in and then click it , second you may decide which plasmid to change . | |
|
Now we enter into the deepest level , we will control E.Coli with our own hand by dragging and dropping some biobricks . Click - Drag - Drop , three simple motion can make a totally different Mr.ecoli . Think about that , thousands of E.Coli is waiting for you to rebuilt them , their willing eyes are looking at you , and expect you could help them make a difference . What are you wating for?! Just do it ! And E.Coli family will appreciate what you do~ | |
|
OK~~Pay attention to what you pick up ! It's very important ! It can directly effect the way our system goes ~ So glare your eyes , use your mind to pick biobrick for our E.Coli . Do you want to see what you've created? Do you want to be god? Click on "simulation" ,and all your dreams will come true ! Show your imagination and talent to the microworld ! To your sincerely E.Coli~~ | |
See Your Result!
| Click Simulate button and see what will happen!
Read more about our modeling idea & modeling algorithm! |
Output
| We use the powerful open source simulator for biochemical networks - COPASI, as the output interface of our software.
It has many functions which will satisfy our needs in most situations. The tasks allowed includes steady-state analysis, time course, parameter scan, etc. The most frequently used function is to perform time course analysis. Users could obtain dynamic curves easily. What's more, it provides convenient way to modify parameters, species and reactions. Users could understand the model's network easily with friendly user interface. |
 "
"