Team:Stockholm/26 August 2010
From 2010.igem.org
Contents |
Hassan
[http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000347190%250D9606.ENSP00000270142%250D9606.ENSP00000368424%250D9606.ENSP00000353940%250D9606.ENSP00000295600%250D9606.ENSP00000241052%250D9606.ENSP00000360157%250D9606.ENSP00000373571%250D9606.ENSP00000267082%250D9606.ENSP00000281043%250D9606.ENSP00000343052%250D9606.ENSP00000364094%250D9606.ENSP00000257880&channel1=off&channel2=off&channel3=off&channel4=off&channel5=off&channel6=on&channel7=on&custom_limit=0&interactive=yes&network_flavor=actions&required_score=700&targetmode=proteins] [http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000347190%250D9606.ENSP00000270142%250D9606.ENSP00000368424%250D9606.ENSP00000353940%250D9606.ENSP00000295600%250D9606.ENSP00000241052%250D9606.ENSP00000360157%250D9606.ENSP00000302777%250D9606.ENSP00000373571%250D9606.ENSP00000354130%250D9606.ENSP00000370171%250D9606.ENSP00000267082%250D9606.ENSP00000281043%250D9606.ENSP00000343052%250D9606.ENSP00000364094%250D9606.ENSP00000257880&channel1=off&channel2=off&channel3=off&channel4=off&channel5=on&channel6=on&channel7=on&custom_limit=0&interactive=yes&network_flavor=actions&required_score=700&targetmode=proteins]
Andreas
Assembly of SOD/yCCS⋅His constructs into pSB1K3
Sequencing results
From 23/8 BLAST results of
pSB1K3.SOD⋅His 1 (pK-SH1)
pSB1K3.SOD⋅His 2 (pK-SH2)
pSB1K3.yCCS⋅His 2 (pK-yH2)
pSB1K3.yCCS⋅His 3 (pK-yH3)
sequences revealed that no His tag was present. In fact, although SOD and yCCS were digested with AgeI for the assembly, the insertion found between SpeI and PstI had not been removed, indicating that SOD and yCCS hadn't even been properly digested; this makes it even more puzzling how the picked clones have been to grow on Km 50 plates, as the SOD and yCCS source plasmid was pSB1C3.
The only reasonable explanation is that SOD and yCCS have been digested only by EcoRI. After enzyme inactivation, however, PstI (which cannot be heat-inactivated) has digested the genes, making them complementary for insertion into pSB1K3 target vector.
Troubleshooting
Two controls were prepared to identify the problems with SOD/yCCS⋅His cloning:
- Gel verification of all samples digested with AgeI and EcoRI
- Dig. pSB1K3.yCCS E+A (yCCS)
- Dig. pSB1K3.SOD E+A (SOD)
- Dig. pMA.His E+A (pMA)
- Plating of sequenced clones onto both Cm 25, Km 50 and Amp 100 plates, to verify the vector resistance.
- pSB1K3.SOD⋅His 1
- pSB1K3.SOD⋅His 2
- pSB1K3.His⋅SOD 1
- pSB1K3.His⋅SOD 3
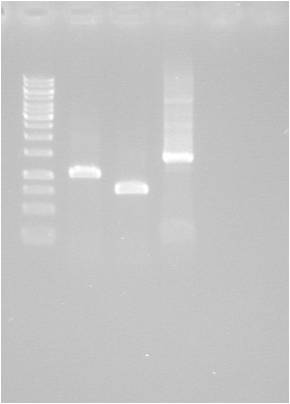
Gel verification
1 % agarose, 110 V, 40 min
Results
Fairly poor digestion of yCCS and SOD. Unclear for pMA.His, since the His fragment is too small to appear on the gel.
Transfer of RFP cassette into pEX
Transformation results
From 25/8
Good yield of both red and white colonies. Four red clones picked for colony PCR verification. PCR performed by Mimmi.
Tranfer of MITF into pSB1C3
Continued from 25/8
MITF taken over by Mimmi.
Amplification of N-CPPs
PCRs prepared from N-CPP cluster plasmid to isolate our three N-CPPs.
- N-Tra10
- Fwd: pSB-VF2
- Rev: pSB-VR
- N-TAT
- Fwd: pEXf
- Rev: pEXr
- N-LMWP
- Fwd: pGexf
- Rev: pGexr
Procedures according to the standard colony PCR protocol. Elongation time 0:45.
Mimmi
His.SOD/His.yCCS
plasmid mini rep
- Transfer cell culture to two 12ml falcon tubes
- Spinn down the cells at 3000rpm for 10m
- Follow the E.Z.N.A mini prep kit 1 protocol
- Wash two times with DNA Wash buffer
- Eluate in 50µl sH2O
- DNA concentration
His.SOD His.yCCS ~160ng/µl ~230ng/µl
MITF-M
Gel
| well | sample |
|---|---|
| 1 | ladder |
| 2 | MITF 1 |
| 3 | MITF 2 |
| 4 | Positive control |
New transformation
- Follow the original protocol!
- Let the plate dry in the incubator before use
pEX.RFP
verification PCR
| Mix | (µl) | X5 | Primer | Conditions | ||||
|---|---|---|---|---|---|---|---|---|
| sH2O | 22.5 | 112.5 | pEX_VF | time | °C | |||
| F primer | 1 | 5 | pEX_VR | 2m | 94 | |||
| R primer | 1 | 5 | 30s | 94 | ) | |||
| DNA | 0.5 | 5X0.5 | 30s | 60 | > 30 cycles | |||
| tot | 25µl | 2m | 72 | ) | ||||
| 10m | 72 | |||||||
| oo | 10 | |||||||
CPPn
quick transformation
- Keep on ice 5m
- Heat shock in 42°C for 30s
- Cool down on ice
- Plate on Amp-plate (dried in the incbator)
 |

|
 |

|
 |

|

|

|
 "
"