Team:Groningen/Expression
From 2010.igem.org
Subtilin induced expression of chaplins
The Biofilm forming capacity of Bacillus subtilis makes it a good host for our application. In addition, B. subtilis is known for its ability to produce and export large amounts of protein at high cell densities. However, despite its track record as an efficient production organism and the fact that both B. subtilis and Streptomyces coelicolor are gram-positive bacteria, it is not certain wether chaplins can be heterologously expressed in B. subtilis. Improper folding, unsuccessful export, or even the very nature of the chaplins, could still lead to hampered expression. We took several steps to ensure optimal expression. The Coding sequences of the chaplins were codon optimized for B. subtilis and synthesized. We placed a ribosome binding site in front of the coding sequences that is known to work well in B. subtilis, and flanked these constructs with the biobrick prefix and suffix.
SURE system
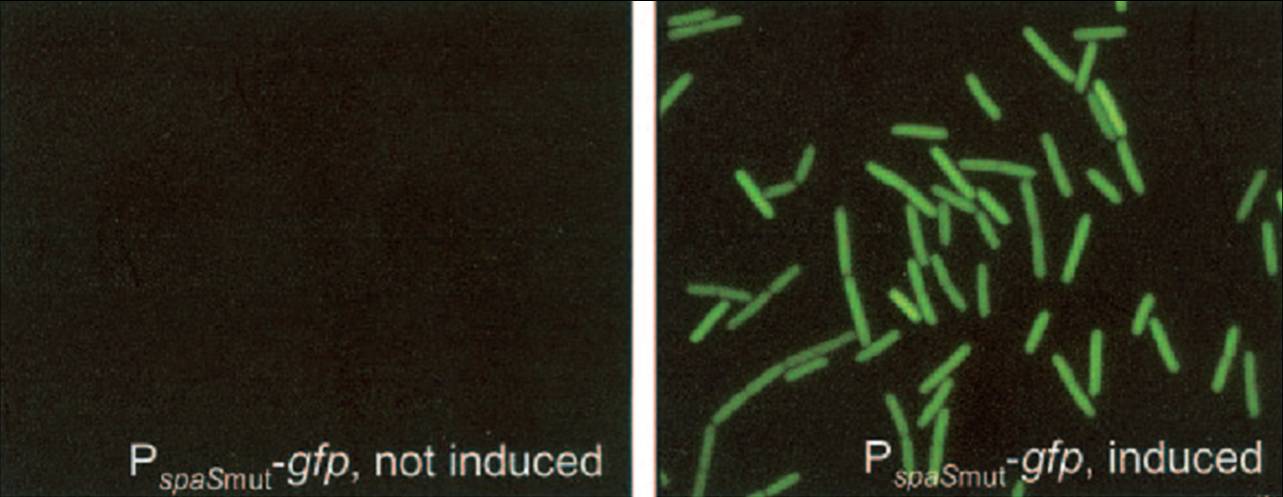
Because it is uncertain how chaplin expression will affect B. subtilis, the initial expression attempts were performed with the stringently controlled, subtilin-regulated gene expression (SURE) system (Bongers et al, 2005). This system uses the subtilin sensing machinery present in a strain of B. subtilis that autoinduces the production of more of the [http://en.wikipedia.org/wiki/Lantibiotics lantibiotic] subtilin. The subtilin sensor histidine kinase SpaK phosphorylates the response regulator SpaR, which can then bind to so-called spa boxes in the promoter regions of genes involved in subtilin biosynthesis (Kleerebezem et al, 2004). In the SURE system, a B. subtilis strain naturally lacking the subtilin biosynthesis genes has the spaRK genes introduced into its genome. A plasmid carrying a spa box promoter that is transformed to this strain can then drive the expression of proteins upon subtilin induction of SpaRK signalling. For our project, we have adapted this system to make it BioBrick compatible with any part that includes an RBS followed by a protein coding sequence. Chaplins
Timed expression of chaplins using biofilm promoters
An important question is which promoter we should use to control the chaplin expression. We assume that an ideal promoter would not be active until the biofilm has formed because the expression of hydrophobic proteins might influence the formation of it. Two promoters where found that are active in biofilms but not during normal growth.
srfA
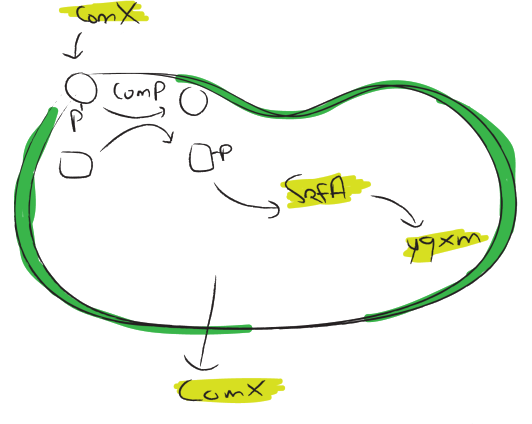
The [http://dbtbs.hgc.jp/COG/prom/srfAA-srfAB-comS-srfAC-srfAD.html srfA operon] has been reported to be important for natural competence and sporulation in Bacillus Subtilis. All these activities occur in biofilms, the promoter is not active until the end of exponential growth. It is controlled by the ComXPA quorum sensing system and hence active in states of high cell densities. Therefore the srfA promoter would be suitable for chaplin expression. Two different lengths of the srfA promoter where chosen due to uncertainties concerning the region between the response element ant the transcription start side of the srfAA protein. In the original promoter this region is unusually long, by shortening it 190bp’s we hope to achieve a higher transcription efficiency. So we came up with two different promoters, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305008 original] one and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305007 shortened] one.
yqxM
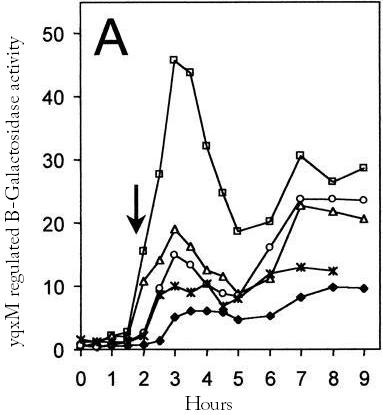
The [http://dbtbs.hgc.jp/COG/prom/yqxM-sipW-tasA.html yqxM-sipW-tasA] operon is controlled by the yqxM promoter. It is needed for biofilm formation because tasA is a key protein of the extracellular matrix. The promotor gets activated via a cascade of other regulatory elements, including srfA, in response to quorum sensing. Since the caplins should work in a similar way to tasA we think the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305006 yqxM] promoter would be very suitable for chaplin expression during the stationary phase.
References
Bongers RS, Veening JW, Van Wieringen M, Kuipers OP, and Kleerebezem M. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl Environ Microbiol 2005 Dec; 71(12) 8818-24. pmid:16332878
Kleerebezem, M., R. Bongers, G. Rutten, W. M. de Vos, and O. P. Kuipers. 2004. Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. Peptides 25:1415–1424.
Stöver AG, Driks A. Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. [http://jb.asm.org/cgi/content/short/181/17/5476 J. Bacteriol. Sept. 1999, p. 5476-5481, Vol. 181, No. 17]
Nakano MM, Xia LA, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. PMID 208261
Frances Chu, Daniel B. Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter and Richard Losicka, A Novel Regulatory Protein Governing Biofilm Formation in Bacillus subtilis PMID 2430766
Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. PMID 1251593
 "
"