Team:EPF Lausanne/Project asaia
From 2010.igem.org


Contents |
Asaia: A new chassis to fight mosquito-borne diseases
One of the main goals of our project is to establish Asaia as a new chassis. We have done a lot of characterization experiments so that other teams will be able to quickly and efficiently engineer new and more potent Asaia strains.
Among the bacteria that naturally live in the mosquito's intestinal tract, we chose the bacteria Asaia because of its high rate of transmission between mosquitoes. Indeed, it seems that Asaia is transmitted horizontally (from one mosquito to another) but also vertically (from parent to offspring). These properties of Asaia make it a really good candidate for our project. Indeed, a lot of mosquitoes would have to be infected with our engineered Asaia before any significant reductions of malaria transmission could occur.
We believe it could be a really good instrument in the fight against malaria and other mosquito-borne diseases.
In this page we want to summarize the main properties coming out from our experiments.
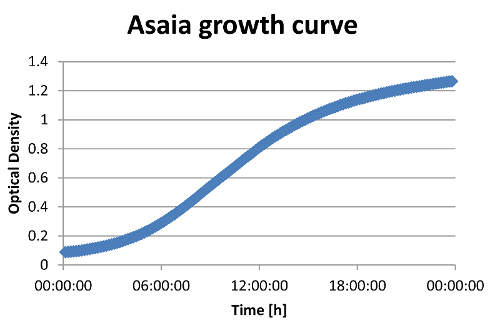
Doubling Time
We measured the growth curve of Asaia at different pH (pH2-pH7) and at 30°C and 37°C to figure out the optimal growth conditions. We found that the optimal Temperature is 30°C, and the optimal pH is 5. At this conditions we measured a doubling time of 2h40min.
Antibiotic resistance
We characterized Asaia's properties against various antibiotics. We had noticed previously that Asaia is naturally resistant to Ampicillin (Amp) and Chloroamphenicol (Cm) to the standard concentrations for E. coli. We tested the behaviour of Asaia to 7 others antibiotic like Tetracyclin (Tet) and Kanamycin (Kan) at various concentrations.
Compatibility with E. coli
We noticed that the Origin of replication of Asaia is compatible with E. coli, but the opposite is not true. The origin of replication of E. coli isn't compatible with Asaia. Thus, we had to add Asaia's origin of replication to all our parts so they would function in both Asaia and E. coli

 "
"