Team:Groningen/28 June 2010
From 2010.igem.org
Week 26
Arend Jan
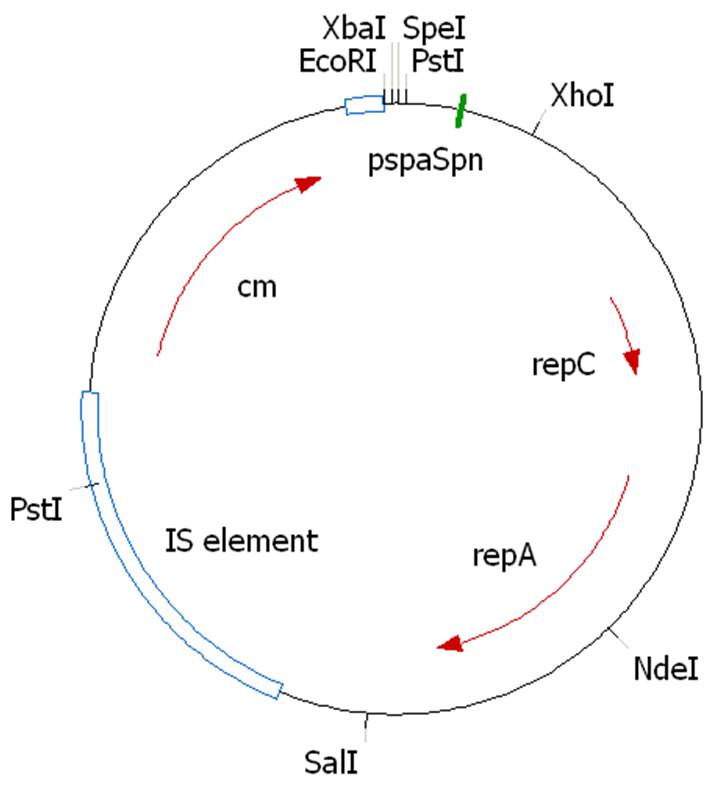
The construct was sent for sequencing (ServiceXS) to sequence the region where the Biobrick sites were inserted, and to sequence the unknown sequence containing the PstI site, using the following primers.
3'RepA-seq-for: ATTGAGAGGAGGGATTATTG
5'Cm-seq-rev: GAGTTTATCACCCTTGTC
3'Cm-seq-for: TTCAGGAATTGTCAGATAGG
The BioBricks were inserted correctly. The unknown sequence was also sequenced correctly. Blasting it revealed that the sequence represents an insertion sequence element containing the IS1 proteins InsA and InsB. It appears this IS element has jumped onto our plasmid.
The PstI site prevents us from inserting two biobricks at once into our expression plasmid. As our synthesized chaplin genes have arrived we will start inserting single genes into the plasmid, not using PstI.
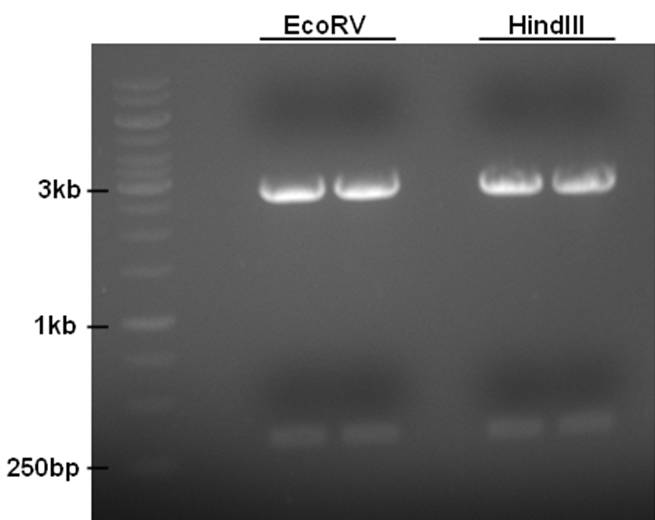
ChpE and ChpH are and the same Mr. Gene construct (pMASK-EH) and are flanked by EcoRV, and HindIII, respectively.
- 10ul plasmid - 1.5ul buffer R - 0.5ul EcoRV/HindIII - 3ul MQ
Fragments were isolated from gel and purified using Roche High Pure PCR Product Purification Kit. The fragments need to be cut with EcoRI and SpeI to ligate them in the expression backbone. However, the DNA concentration after purification was to low to continue. Repeat with more plasmid.
 "
"