Team:Harvard/allergy/notebook
From 2010.igem.org

notebook calendar
06-14-2010 [ top ]
- Task #1
- Task #2
- Task #3
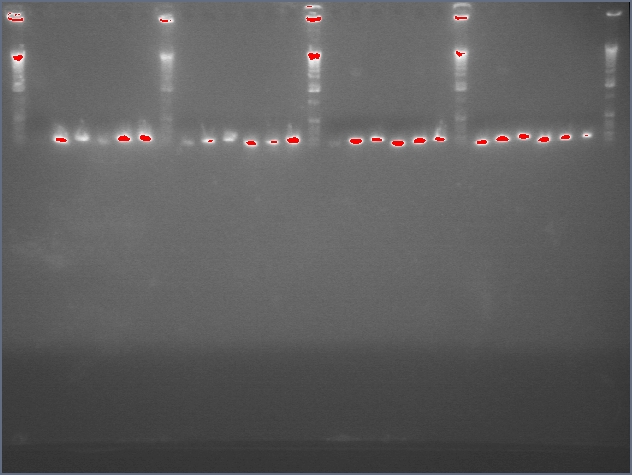
Image of gel, photo of your work, diagram, etc.
Results/ Conclusions
- We successfully cloned x, y, z
- Next step is to ligate 1, 2, 3
06-15-2010 [ top ]
06-16-2010 [ top ]
06-17-2010 [ top ]
06-18-2010 [ top ]
06-21-2010 [ top ]
06-22-2010 [ top ]
06-23-2010 [ top ]
06-24-2010 [ top ]
06-25-2010 [ top ]
06-28-2010 [ top ]
06-29-2010 [ top ]
06-30-2010 [ top ]
07-01-2010 [ top ]
07-02-2010 [ top ]
07-05-2010 [ top ]
07-06-2010 [ top ]
07-07-2010 [ top ]
07-08-2010 [ top ]
07-09-2010 [ top ]
07-12-2010 [ top ]
07-13-2010 [ top ]
07-14-2010 [ top ]
07-15-2010 [ top ]
07-16-2010 [ top ]
07-19-2010 [ top ]
07-20-2010 [ top ]
07-21-2010 [ top ]
07-22-2010 [ top ]
07-23-2010 [ top ]
07-26-2010 [ top ]
07-27-2010 [ top ]
07-28-2010 [ top ]
07-29-2010 [ top ]
07-30-2010 [ top ]
08-02-2010 [ top ]
08-03-2010 [ top ]
- Grew up cultures of completed ihpRNA constructs (Bet, LTP, Ger) in pORE expression vector
- amiRNA PCR
- Will look at results of PCR tommorrow
08-04-2010 [ top ]
- amiRNA PCR appears to have worked at every Tm we tried:
- Digested V9/V10 to insert our constructs into
- Realized that we hadn't gel purified our ihpRNA inserts
- Gel purification of inserts (entire ihpRNA parts)
Gel 1: Ladder, 9, 11c1, 11c2, ladder, 25c1, 28c1, 28c2, 36c1, 36c2, ladder Gel 2: Ladder, 37c1, 37 c2, 38c1, 38c2, ladder
08-05-2010 [ top ]
08-06-2010 [ top ]
08-09-2010 [ top ]
08-10-2010 [ top ]
08-11-2010 [ top ]
08-12-2010 [ top ]
08-13-2010 [ top ]
Procedures
Gel Electrophoresis
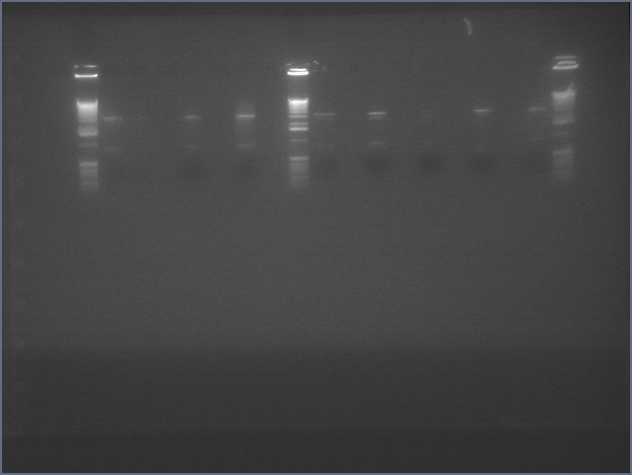
Today, we loaded gel slots to run our 6 DNA samples against a 1kb ladder. The order of our slots were: ladder, ladder, sense 1, sense 1, sense 2, sense 2, sense 3, sense 3, antisense 1, antisense 1, antisense 2, antisense 2, antisense3, and antisense 3. We had to load two slots because there was more than 50 microliters of DNA and it would not fit into one slot. Since we are hoping to collect and purify the DNA after the electrophoresis, we want to run as much of the sample as possible. We also loaded two ladders. We ran the gel for 30 minutes at 100 volts. Our goal is to have the majority of the amplified DNA to be 300bps long.
Thirty minutes later, we examined our gel under UV light. All of the DNA were smaller than 100bp, suggesting that the Phusion Polymerase did not amplify the correct DNA segment. Our sample looks like a primer dimer.
Results
If our Phusion Polymerase had worked, then we would have continued to extract the correctly formed DNA segments, performed a restriction digest and cut the PCR products into a B0120 biobrick with Xba1 and Pst1, ligate, and transform into E. coli. However, since we do not have the correct DNA, we cannot move forward.
Errors
There are several reasons for why our gel did not work. The most likely reasons is that the RNA extraction did not work properly. We used strawberry fruit as our sample for RNA extraction. Because RNA is constantly being degraded by RNAases, the integrity of our RNA may have been compromised before the addition of the denaturant.We will re-extract DNA from our plant samples, now including arabidopsis, probably tomorrow.
Another potential error could have occurred during the Phusion Polymerase. We set our annealing temperature at 72°C, which could have been too high for three of our four primers. We'll rerun our PCR with a gradient PCR of annealing temperatures to see if we can get a better result. The amount of DNA added by template was also much higher than the optimal 50~250 nanograms per 50 micrograms of PCR reactants. We will dilute the DNA templates to 100 micromolar and use one microliter per reaction so that there are approximately 100 nanograms of DNA per reaction.
The Next Step...
Corrections
Redo gel electrophoresis with all three concentrations of Antisense and Sense, each at a range of temperatures, from one to three degrees Celsius above the lower annealing temperature of the primers. This will give three tubes of each of three concentrations of both Sense and Antisense, for a total of 18 tubes. Run gel for 30 minutes at 100 volts with a 100kb ladder.
Corrections were unsuccessful. We are going to go further back and do more research on the strawberry allergen and see if we can obtain a better RNA and DNA sample.
Strawberries are also non-climacteric fruits, which means that they cease to respire after being picked. Our sample strawberries were picked from a grocery store, so whatever mRNA responsible for ripening would have degraded after picking. Climactic fruit, like tomatoes and apples, continue to metabolize sugars and ripen after being picked. The mRNA we were hoping to obtain is related to the ripening process of strawberries, but since our strawberries were not fresh, it is possible that all of the mRNA responsible for coding Fraa1 had degraded before our RNA extraction was performed.
Further procedures If the gel electrophoresis is successful and the pieces are 300bps, then we will perform:
- Gel Extraction - remove the correct Antisense and Sense DNA segments
- Restriction Digest - cut the DNA segments with Eco1 and Xba1 (keeping A and S separate)
- Ligation - links sense, intron, and antisense into a V0120 plasmid (Biobrick)
- E. coli Transformation - insert the plasmid into bacteria
 "
"