Team:USTC Software/Features
From 2010.igem.org
Contents |
Fun and Function
MoDeL: Modeling Database Language
Bring Biological Modeling to the Next Level
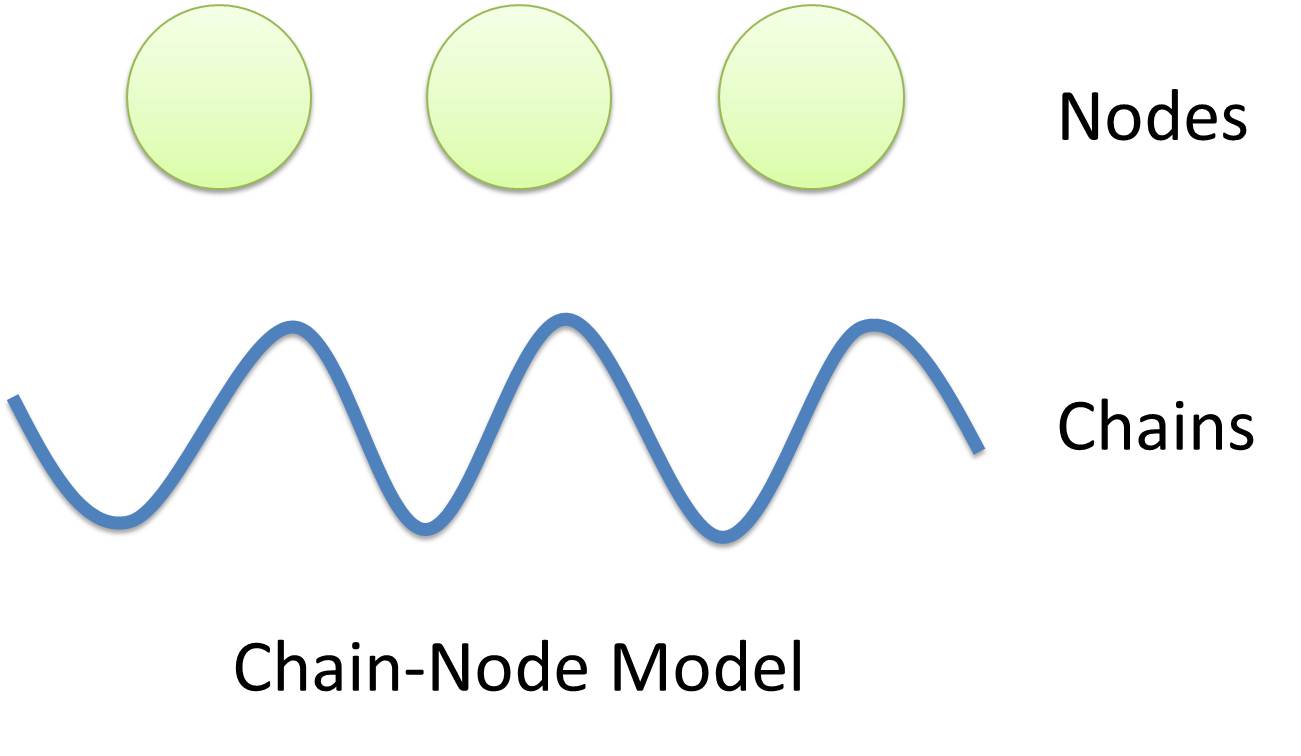
| Chain-Node Model (Figure. 1) is a brand new Complex Modeling Concept incorporating its detailed structure description with universal applicability. Instead of treating complex as a whole while ignoring their basic composition and structure, Chain-Node Model view complex as a construction of it basic Parts. Just as its name implies, our model includes two components: Chain and Node. As a correspondence to natural polymer chains, each Chain consists of an arrangement of its basic unit, Part, whose concept has been greatly extended and includes but not limited to Biobrick Parts. The Node component is not a natural correspondence but an abstract concept to describe binding states of two or more parts: each binding will create a Node. The abstract nodes may continue to bind with other parts or nodes to form a tree structure. However, parts or nodes in bound states are not allowed to bind again. With help of chains and nodes, it is possible to model any complex with arbitrary architecture. Simple and inaccurate modeling of biological process could not keep pace with the development of synthetic biology and undoubtedly, our Chain-Node model provides a possible solution to the imbalance. | |
| A simple example, tetR dimer, is shown to illustrate our simple modeling idea (做一个tetR2的模型放在右边做为配图,否则右边太空了). It has two chains with each containing only one part, tetR. Dimerization of tetR will create a node to indicate the bound state of two parts. To explain more clearly, bound parts are also considered as nodes so that in this example, all nodes are organized in a tree structure, which includes two children (leaf) nodes and one parent node. We will conform to this convention in our wiki.
| |
| To know more, users are suggested to read this One-Minute Introduction to have an intuitive idea and deeper understanding of our modeling system. | |
Modeling with Templates
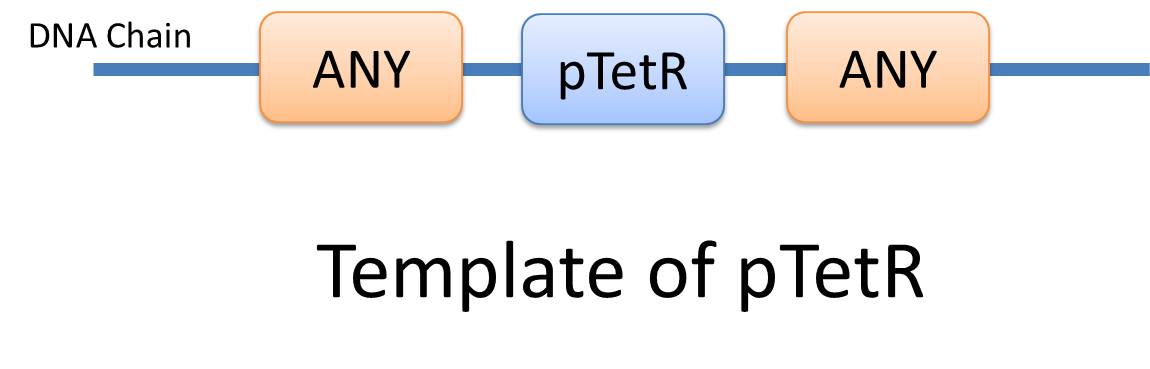
| Similar reactions happen for similar reasons. For example, the repression of pTetR promoter by TetR dimer can happen on many different DNA molecules with pTetR, regardless of what other biobricks is constructed on the DNA. With this thought, we add the part of Substituent, which can replace inactive parts of Species in Reactions, making it more general, and actually turning it into a reaction template. Having substituent, a pTetR TetR binding reaction is re-interpreted as the binding reaction of ANY DNA with pTetR(Figure 2) with ANY protein with TetR-dimer binding site. Modeling with templates allows us to describe reactions of new complex even without rewrite the reactions and species in database. |
Automatic Modeling Database Language
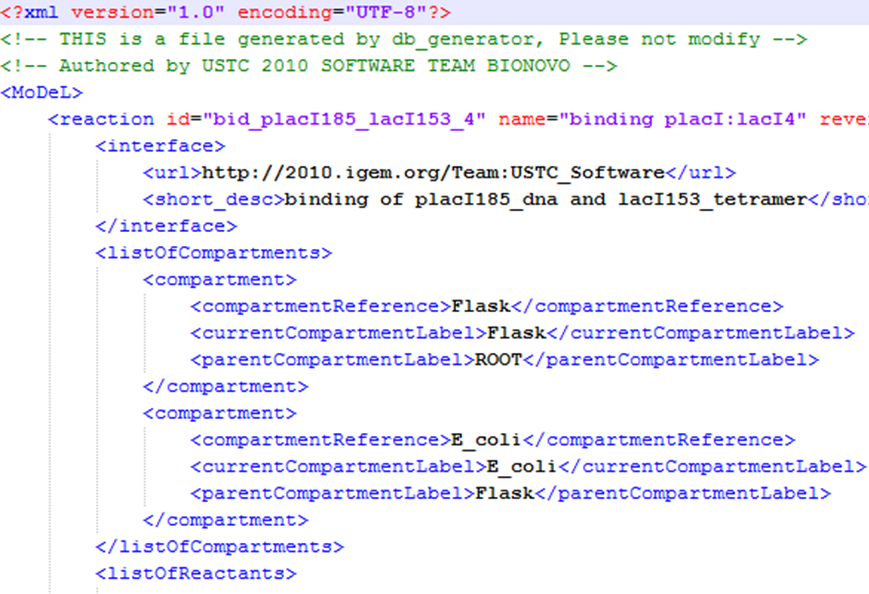
| We use a database to store all the information we need in modeling. In order to realize automatic modeling, we construct the database in unified format and make it machine-readable. Every component of database has its specified attributes and values, which makes the format of the database a unique yet standard database language. We call it MoDeL: Modeling Database Language by picking out characters from three words. MoDeL is based on XML language, which makes it flexible and extensible. For more specifications of MoDeL, click here. | |
 "
"