Team:Freiburg Bioware/testpage
From 2010.igem.org

76.Labortag 01.08.2010
Picking clones of pSB1C3_CMV
Investigator: Bea
Comments: Trafo was performed friday, and trafo plate were incubated over night. Clones need to be picked in order to perform Mini-Prep
- Bacterial strain used: XL1-B
- Two clones were picked of trafo plate
- Inoculating of 10 mL DYT medium containing 10µL chloramphenicol
- Put in 37°C room on rotary shaker
77.Labortag 02.08.2010
Sequenzing of pSB1C3_RFC25_longlinker, pSB1C3_RFC25_SEG, pSB1C3_RFC25_GSAT
Investigator: Jessica
Comments: Linkers (we got from Gerrit) we cloned in the RFC25-standard (pSB1C3_RFC25). Result looks good, linkers are in the vector pSb1C3_RFC25
- pSB1C3_RFC25_longlinker P105 und P106
- pSB1C3_RFC25_SEG P107 und P108
- pSB1C3_RFC25_GSAT P109 und P110
Sequencing of pGA14_Prefix-leftITR
Investigator: Hanna
Eventually GATC partially managed to sequence pGA14_Prefix-left ITR clone 1 and 2.
The sequencing results were better than before but in the ITR region still not evaluable due to the strong secondary structures.
It seemed that, despite of successful insertion of the RFC10-Prefix, something went wrong because there were 2 bases missing in the NotI restriction site. But by having a closer look, it became obvious that the Geneious misinterpreted the chromatogram and overlaid the peaks of the missing bases.

For the first time sequencing of parts of the 3' end worked: The remaining NotI restriction site, which was already eliminated this week and also the "backbone-PstI" can be seen:

Sequencing of very GC-rich regions :) :) :)

Sequencing of clone 4 delivered that the ITR and the RFC10-Prefix were not inserted in the vector. The RFC25 standard of the multiple cloning site is still in the plasmid:

Therefore clone 4 was dicarded from the plasmid- and glycerol stock.
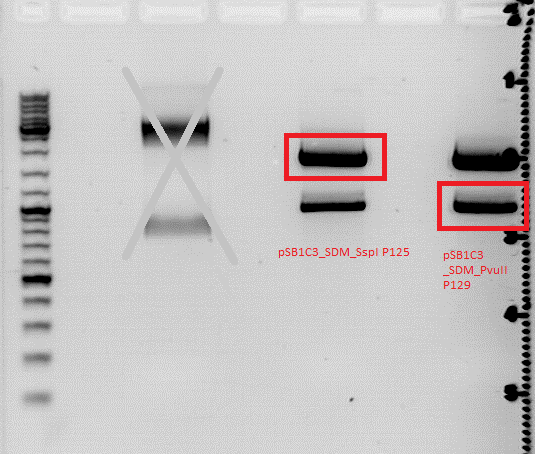
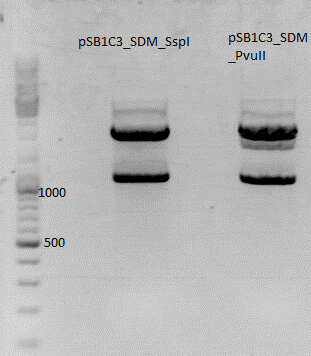
Repetition of test digestion pSB1C3_CFP SDM SspI and PvuII
Investigator: Jessica
- buffer used: 2; Restriction-enzymes used: Enzyme 1: SspI ; Enzyme 2: PvuII
- Plasmids (all: ~ 700 ng/µL):
- pSB1C3_CFP P51.1
- pSB1C3_SDM_SspI P125
- pSB1C3_SDM_PvuII P129
- pSB1C3_SDM_PvuII P131
| Components | Mastermix | P125/µL | P51.1.1/µL | P129/µL | P131/µL | P51.1.2/µL |
| DNA | - | 2,4 | 2,3 | 1,6 | 5,3 | 5,3 |
| BSA (10x) | 6 | 1 | 1 | 1 | 1 | 1 |
| Buffer 2 (10x) | 6 | 1 | 1 | 1 | 12 | 1 |
| Enzyme 1 SspI (68) | - | 1 | 1 | - | - | - |
| Enzyme 2 PvuII (50) | - | - | - | 1 | 1 | 1 |
| H2O | - | 4,6 | 4,7 | 5,4 | 1,7 | 1,7 |
| Total volume | 12 | 10 | 10 | 10 | 10 | 10 |
- Incubation: 65 minutes
Agarosegel
0.45 g Agarose, 50 ml TEB (0,5 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
- Marker: GeneRuler ladder mix (6x)
| Marker | P125 | P51.1 SspI | - | P129 | P131 | P51.1 PvuII | |
|---|---|---|---|---|---|---|---|
| Lane | 7 µL | 10 µL | 10 µL | - | 10 µL | 10 µL | 10 µL |
- P125 isn't cut, therefore you see the bands for supercoiled, nicked and relaxed
- P51.1 is cut once, therefore you can see a faster running band
- P129 and P131 should be cut once because of the PvuII in the CFP but there is another band we can't identify
- P51.1 should be cut two times but there is also another unidentified band
- perhaps there is another PvuII in the vector we can't find (???)
Comments: SDM of SspI is ready! SDM of PvuII looks confusing because of the lower band. P 129 will be sequenced but this doesn't solve this problem. there has to be one more rs we don't know, perhaps. we will check it...anywise
Mini-Prep and Test digestion of pSB1C3_CMV
Investigator: Kerstin
Nanodrop
- pSB1C3_CMV clone1: 157,6 ng/µl P145
- pSB1C3_CMV clone2: 155,6 ng/µl P146
Test digestion 900ng DNA
| components | P145 /µl | P146 /µl |
| DNA | 5,7 | 5,8 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4 (10x) | 0,5 | 0,5 |
| Enzyme 1: XbaI | 0,5 | 1,5 |
| Enzyme 2: PstI | 0,5 | 0,5 |
| H2O | 5,3 | 5,2 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 15 |
agarose gel: 1,5%, digestion: 90 minutes, 37°C
P145 is sent for sequencing with Reverse Primer (VR2)
Comments:Results look good. CMV has the expected size (~ 662bp)
</p>
Sent for sequencing
Investigator: Jessica
Hanna
- P145 left ITR
- Primer: GATC_std_pQE-FP and GATC_std_m13-FP
- P150 right ITR
- Primer: GATC_std_pQE-FP and GATC_std_m13-FP
Anissa
- P136 pSB1C3_hgh
- Primer: O51_Reverse Primer (VR2)
- P139 pAAV_BamHI
- Primer: O36_Rep_1250_rev
- P143 pAAV_RC_SalI
- Primer: O53_Rep_1250_for
- P141 pGL3_QTERT
- Primer: GATC_std_pTal-FP
- P133 pSB1C3_betaGlobin
- Primer: O51_ReversePrimer (VR2)
- P145 pSB1C3_CMV
- Primer: O51_ReversePrimer (VR2)
Jessica
- P129 pSB1C3_SDM_PvuII
- Primer: GATC_std_pTeSp-2
Preparation of competent E.coli
Investigator: Jessica
- 15ml DYT was prepared with 15µl tetracycline and inoculate with XL1B
- 10ml DYT w/o antibiotics was prepared and inoculate with BL21 (just for glycerol stock, no competent cells)
both were incubate over night in 37°C room
Last Mini-Prep and test digestion of ITRs
Investigator: Hanna
Plasmid Mini-Prep
- new vector name: pGA14_RFC10_leftITR and pGA14_RFC10_rightITR
Glycerol Stocks
pGA14_RFC10_leftITR
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1blue | XL1blue | XL1blue |
| Plasmidname | pGA14_RFC10_leftITR | pGA14_RFC10_leftITR | pGA14_RFC10_leftITR |
| Date | 2.8.10 | 2.8.10 | 2.8.10 |
| given glycerol-stock no. | B115 | B116 | B117 |
| given plasmid no. | P147 | P148 | P149 |
pGA14_RFC10_rightITR
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1blue | XL1blue | XL1blue |
| Plasmidname | pGA14_RFC10_rightITR | pGA14_RFC10_rightITR | pGA14_RFC10_rightITR |
| Date | 2.8.10 | 2.8.10 | 2.8.10 |
| given glycerol-stock no. | B112 | B113 | B114 |
| given plasmid no. | P150 | P151 | P152 |
Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 XbaI ; Enzyme 2 SpeI
- Plasmid: pGA14_RFC10_leftITR:
- Given Plasmid-Number: P147; DNA concentration: 240.22 ng/µL ;
- Given Plasmid-Number: P148; DNA concentration: 268.97 ng/µL ;
- Given Plasmid-Number: P149; DNA concentration: 234.16 ng/µL ;
- Plasmid: pGA14_RFC10_rightITR:
- Given Plasmid-Number: P150; DNA concentration: 221.71 ng/µL ;
- Given Plasmid-Number: P151; DNA concentration: 191.13 ng/µL ;
- Given Plasmid-Number: P152; DNA concentration: 215.14 ng/µL ;
| Components | P147 Volume/µL | P150 Volume/µL |
| DNA | 4.2 | 4.5 |
| BSA (10x) | 1 | 1 |
| Buffer no. 4 (10x) | 1 | 1 |
| Enzyme 1 XbaI | 0.5 | 0.5 |
| Enzyme 2 SpeI | 0.5 | 0.5 |
| H2O | 2.8 | 2.5 |
| Total volume | P147 | P150 |
- Incubation: 1 h
Agarose-Gel:
0.83 g Agarose, 53 mL TBE (1.57%), 3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P147 | 10 µl | 2 µl | 157 bp | 2902 bp |
| P150 | 10 µl | 2 µl | 156 bp | 2903 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P147 /µL | Sample P150 /µL | |
|---|---|---|---|
| Lane | 6.5 | 12 | 12 |
Test digestion delivered positive results for both ITRs: The expected fragment sizes (156 resp. 157 bp) could be detected in the referring range between the 100 and 200 bp marker nucleotides:

Comment: In order to verify the results, P147 and P150 were sent for sequencing. Because sequencing never worked before, they will be sequenced under special conditions this time.
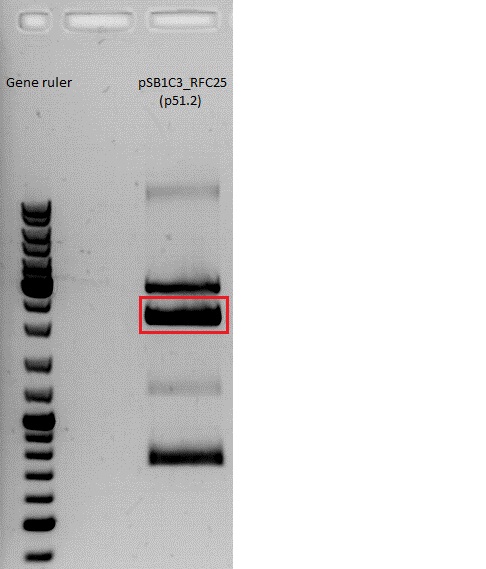
BioBrick production: mGMK in pSB1C3
Investigator: Stefan
Backbone taken from:
- pSB1C3_CFP (P51.2): 151,1 ng/µl
Insert taken from:
- pAAV_RFC25_mGMK (P100): 411,3 ng/µl
| components | pSB1C3_CFP(P51.2) /µl | pAAV_RFC25_mGMK (P100) /µl |
| DNA | 8 | 5 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| XbaI | 1 | 1 |
| AgeI | 1 | 1 |
| H2O | 6 | 9 |
| Total volume | 20 | 20 |
- 1% Agarose gel
- 3 µl Gelred
- 7 µl DNA-Ladder-Mix
- 115 Volt, running time: 50 minutes
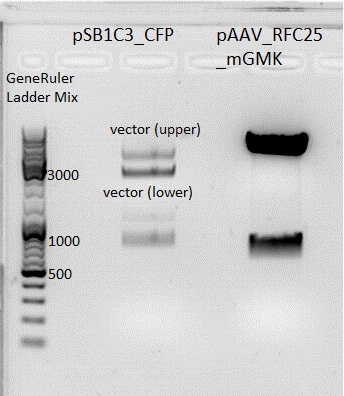
Afer 50 minutes a picture (see below) was taken. Because there was no clear pattern (two bands) for the pSB1C3 backbone, the band of the insert mGMK was cut out and the gel ran another 20 minutes. After that, there were still two bands. Both were cut out and it was continued with two possible backbones. They were labeled vector (lower) and vector (upper).
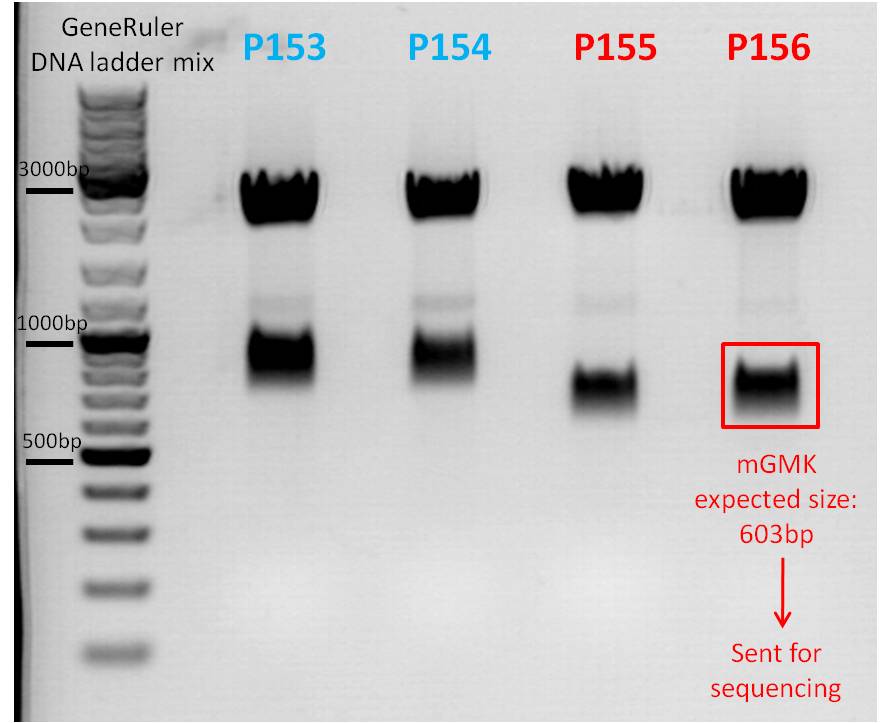
| Sample/µl | Expected size/bp |
| Vector pSB1C3 | 2082 |
| Insert mGMK | 603 |
- weight of insert mGMK gel extract: 0,17 g
- weight of vector (lower) pSB1C3 gel extract: 0,08 g
- weight of vector (upper) pSB1C3 gel extract: 0,09 g
Nanodrop
- insert mGMK : 4,13 ng/µl
- vector (lower) pSB1C: 5,47 ng/µl
- vector (upper) pSB1C: 0,60 ng/µl => for calculation of ligation approach calculated with 1 ng/µl
Ligation
- vector (lower) : insert - 4,16 µl : 4,84 µl
- vector (upper) : insert - 7,43 µl : 1,57 µl
Transformation
Two approaches were prepared, one for each vector and put in 37 °C room overnight.
Sequencing results of SDM Rep 68 & 78
Investigator: Hanna and Volker
The sequencing results delivered that the PstI restriction sites of Rep 68 and Rep 78 were successfully deleted via side-directed mutageneis:
78.Labortag 03.08.2010
Results of sequencing pSB1C3_CFP_SDM_PvuII
Investigator: Jessica
SDM of PvuII:
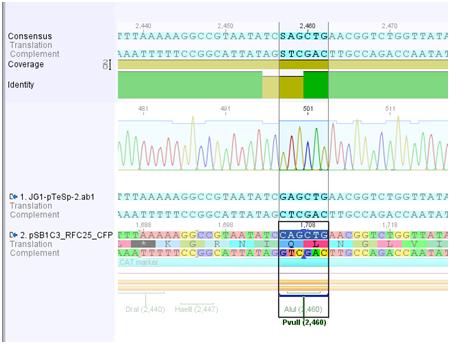
PvuII is succesfully deleted in the vector pSB1C3_RFC25_CFP
Repetition of the quickchange site-directed mutagenesis of pAAV_RC_1.1_SalI
Investigator: Kerstin,Anissa
Comments: Sequenzing revealed no mutagenesis of SalI in pAAV_RC_1.1_SalI, so SDM now will be repeated.
PCR-reaction:
| Volume / µl | ingredients | recommended /µl |
| 2,5 | 10X Pfu Ultra II buffer | 2,5 |
| 4,18 | template (~10 ng) | 4,18 of 1:100 dilution |
| 0,58 | forward primer: O68 | 62,5 ng |
| 0,58 | reverse primer: O69 | 62,5 ng |
| 0,5 | dNTP | 250 µM each dNTP |
| 16,16 | H2O | |
| 0,5 | PfuUltra II fusion (1.25) |
PCR program:
| Rounds | temperature/ °C | Time |
| 1 | 95 | 2 minutes |
| 20 | 95 | 30 seconds |
| 20 | 80,4 | 1 minute |
| 20 | 68 | 7,5 minutes |
Plasmid was transformed into BL21 cells. Clones have to be picked tomorrow.
Comments:NO CLONES, trafo didn't work, because annealing temperature was totally to high! (80,4 °C instead of 55°C) annealing temperature should always be 55°C (in case of troubleshooting temperature can be increased up to MAX. 68°C !)
PCR for Biobrickproduction of Rep 40, 52, 68, 78 and APP
Investigators: Volker, Anna
Aim of the experiment:
We wanted to produce biobricks from the expression plasmids Rep40ex, Rep52 ex, the Rep 68 and 78 in which the PstI at position 310 was silenced and the AAP. This was performed by a PCR with primes that annealed to the ends of the later biobrick but also contained restriction sites that can be cut afterwards and cloned into pSB1C3.
- Plasmids used as template:
Rep_68_ex (p119): c = 470,6 ng/µl
Rep_78_(p122): c = 201,08 ng/µl
Rep_40_(p22.2): c = 673,1 ng/µl
Rep_52_(p23.2): c = 532,22 ng/µl
pAAV-RC containing AAP ORF (p50): c = 378,5 ng/µl
- Primer used:
For Rep_40_ex: Praefix_40_52_ex & Suffix_40_68_ex
For Rep_52_ex: Praefix_40_52_ex & Suffix_52_78_ex
For Rep_68_ex: Praefix_68_78_ex & Suffix_40_68_ex
For Rep_78_ex: Praefix_68_78_ex & Suffix_52_78_ex
For AAP_ex: Praefix_AAP_ex & Suffix_AAP_ex
- PCR:
(was performed following the standard protocol)
| Ingredients | Volume / µl | Rep68 | Rep78 | Rep40 | Rep52 | AAP |
| 5X Phusion HF buffer | 10 | |||||
| 10 mM dNTP mix | 1 | |||||
| forward primer: | 2,5 | |||||
| reverse primer: | 2,5 | |||||
| DNA Template | *** | 4,2 µl | 1 µl | 2,9 µl | 3,8 µl | 5,3 µl |
| Phusion Polymerase | 0,5 | |||||
| H2O | *** | 28,3 µl | 31,5 µl | 29,6 µl | 28,7 µl | 27,2 µl |
| Total volume | 50 |
PCR program:
| PCR Program | temperature/ °C | Time | Rep40 | Rep50 | Rep68 | Rep78 | AAP |
| 1 | 98 | 1min | |||||
| 2 | 98 | 15s | |||||
| 8x | *** | 25s | 63°C | 62°C | 63°C | 62°C | 64°C |
| 3 | 72 | *** | 15s | 18s | 24s | 27s | 10s |
| 4 | 98 | 15s | |||||
| 17x | *** | 25s | 68°C | 66°C | 68°C | 64°C | 68°C |
| 5 | 72 | *** | 15s | 18s | 24s | 27s | 10s |
| 6x | 72 | 5min | |||||
| Hold | 4 |
Five µl of PCR-product were used for an analytical gel to see if the PCR worked.
Comment: The result is that the PCR worked for the longer Rep proteins (68&78) with which a Quick-change was performed to remove the PstI(310) but not with the constructs for the smaller Rep variants (40&52) that were recieved from PD Kleinschmidt. As the longer Rep variants the AAP PCR Reaction also resulted in a PCR-product of the expected size. Unfortunately there were a secondary band of ~90bp for Rep_68_ex. For this reason we performed a praeparative gel and decided to cut the bands of all PCR products.
The bands marked in the gel picture were cut and a gel extraction was performed.
Gel extraction
Gel measurement:
| Sample | Weight | Volume | Concentration |
| Rep_68_ex | 0,1 g | 20 µl | 17,6 ng/µl |
| Rep_78_ex | 0,15g | 20 µl | 80,6 ng/µl |
| AAP_ex | 0,19 g | 20µl | 77,78 ng/µl |
Continuation of preparation of competent E.coli
Investigator: Jessica
preparation of competent XL1blue was finished according to the standard protocol
- aliquots of 60µl (to use for 1 trafo) are stored in -80°C freezer
Sequencing of pSB1C3
Investigator: Jessica
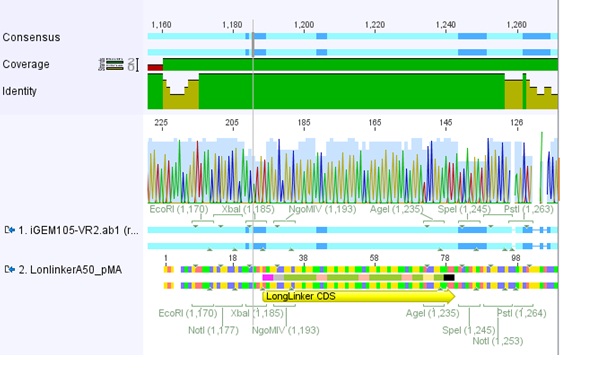
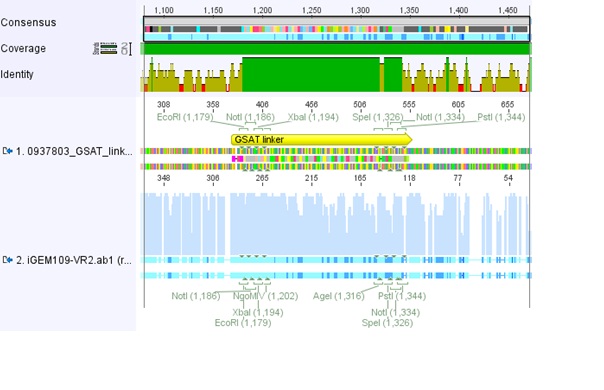
Comments: We sent for sequencing pSB1C3_RFC25_longlinker (P105) to check the strange result of the test digestion of pSB1C3_SDM_PvuII(01.08.10.
pSB1C3_SDM_PvuII is too long for sequencing wherefore we sent pSB1C3_RFC25_longlinker because the longlinker just have 54kb (bp, nicht kb (Volker) (necessary is just the backbone we don't have.
- Primer:
- RESgen-241698
- Reverse primer (VR2)
- o_F1-fw (from Gerrit)
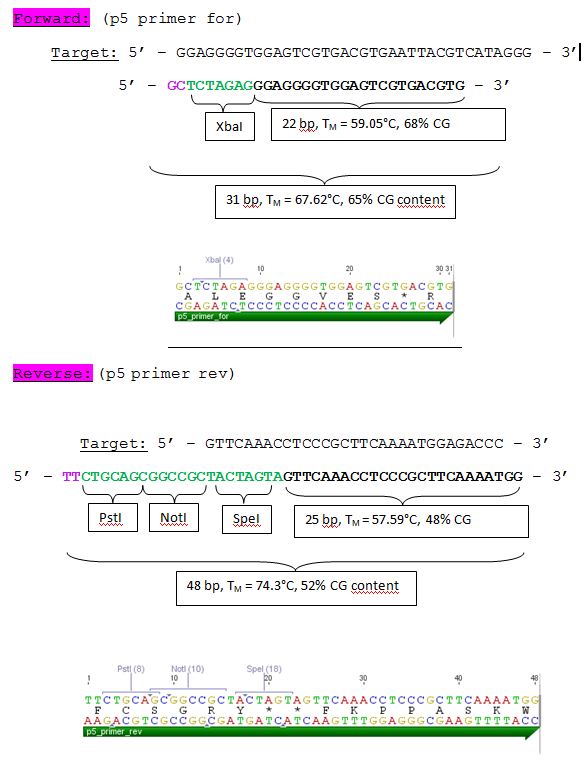
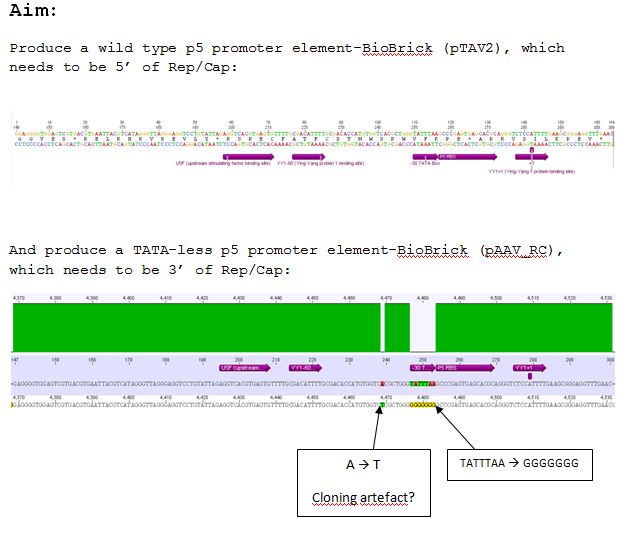
Design of Primer for p5 Promoter WT and TATA-less BioBrick production
Investigator: Hanna
79.Labortag 04.08.2010
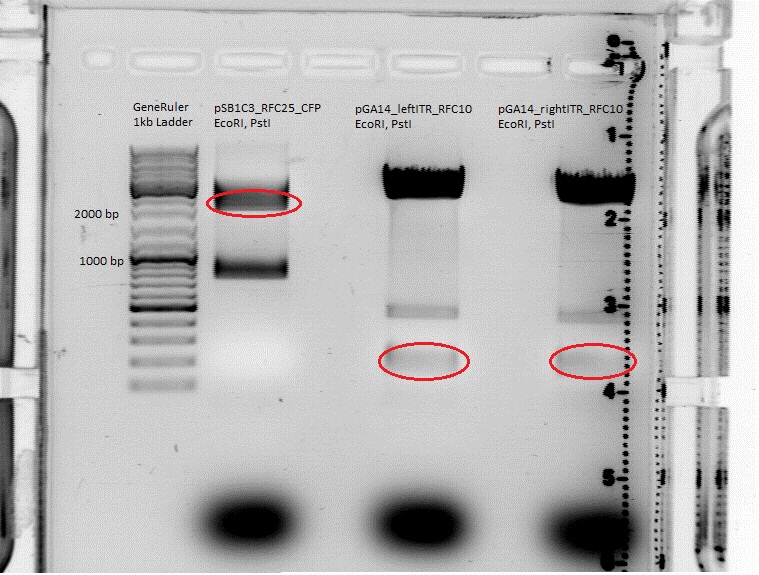
Cloning of right ITR and left ITR into pSB1C3_RFC25_CFP
Intention: Get biobricks ready.
Investigator: Patrick
pGA14_lITR_RFC10 (P147), pGA14_rightITR (P150) and pSB1C3_RFC25_CFP (P51.1) were digested with EcoRI and PstI (Buffer 4) according to the standard protocol. Digestion Time: 80 minutes.
GelRun: 1% Agarose Gel. Expected results (from left to right):
- pSB1C3_RFC25_CFP: about 2100 bp and 800 bp.
- pGA14_leftITR_RFC10: the size of the insert should be 135 bp.
- pGA14_rightITR_RFC10: the size of the insert should be 138bp.
The mutual vector (now without CFP) and inserts (left ITR and right ITR) were cut out. The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pSB1C3_RFC25_CFP: 2,9 ng/µl
- left ITR: 1,4 ng/µl
- right ITR: 2,0 ng/µl
The Quick Ligation was not performed according to the standard protocol:
- left ITR + pSB1C3: 5µl Buffer, 1 µl Quick-Ligase, 2,5 µl left ITR, 1,5 µl pSB1C3_RFC25.
- right ITR + pSB1C3: 5µl Buffer, 1 µl Quick-Ligase, 2,5 µl right ITR, 1,5 µl pSB1C3_RFC25.
Transformation: performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol. The clones will be picked tomorrow.
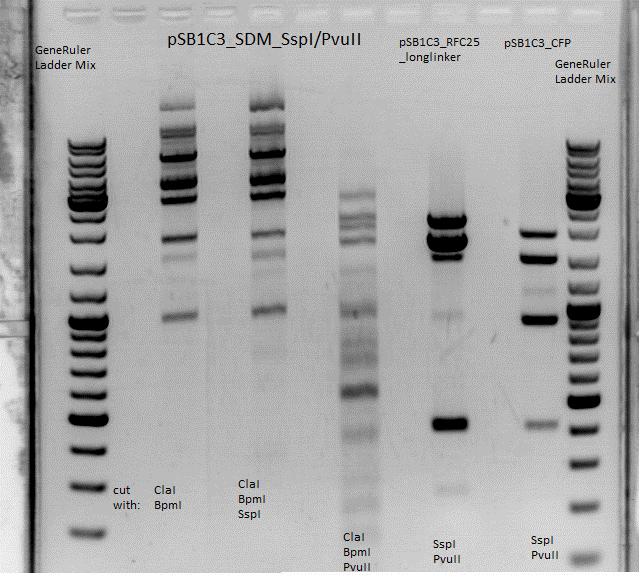
Cloning of SDM SspI and SDM PvuII
Investigator: Jessica
- Vector: name: pSB1C3_SDM_SspI P125
- Insert: name: pSB1C3_SDM_PvuII P129
- new vector name: pSB1c3_SDM_SspI/PvuII P157
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab: 141) BstI ; Enzyme 2 (no.Lab: 144) BpmI
- DNA concentration (vector): 321,9 ng/µl ; DNA concentration (insert): 308,4 µg/µl
| components | volume of pSB1C3_SDM_SspI /µl | volume of pSB1C3_SDm_PvuII /µl |
| DNA | 4,66 | 4,87 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme BstI (no.Lab:141) | 1 | 1 |
| Enzyme BpmI (no.Lab:144) | 1 | 1 |
| H2O | 9,34 | 9,13 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 130 Volt, running time:45
Loading plan for agarose gel:
Marker used: GeneRuler ladder mix (Fermentas)
| Marker | Sample 125, 20µl | Sample 129, 20µl | |
|---|---|---|---|
| Lane | 1 | 3 | 5 |
Results: digesttemperature of BstI is 55°C, plasmid was just digested at 37°C... :( (
2nd Repetition of Quickchange site-directed mutagenesis of pAAV_RC_1.1_SalI
Investigator: Kerstin, Anissa
Comments: Last trafo didn't work. Annealing temperature was to high (80,4°C instead of 55°C)
Two approaches were made (short and long PCR:
PCR-reaction:
| Volume / µl | ingredients | recommended /µl |
| 2,5 | 10X Pfu Ultra II buffer | 2,5 |
| 1,83 | template (~10 ng): p139 (c: 546,97 ng/µl) | 1,83µl of 1:100 dilution |
| 0,58 | forward primer: O68 | 62,5 ng |
| 0,58 | reverse primer: O69 | 62,5 ng |
| 0,5 | dNTP | 250 µM each dNTP |
| 18,51 | H2O | |
| 0,5 | PfuUltra II fusion (1.25) |
PCR program (long):
| Cycles | temperature/ °C | Time |
| 1 | 95 | 2 minutes |
| 20 | 95 | 30 seconds |
| 20 | 55 | 1 minute |
| 20 | 68 | 7,5 minutes |
PCR program (short):
| Cycles | temperature/ °C | Time |
| 1 | 95 | 2 minutes |
| 20 | 95 | 30 seconds |
| 20 | 55 | 1 minute |
| 20 | 68 | 4 minutes |
Plasmids were transformed into BL21 cells. Clones have to be picked tomorrow.
Mini-Prep of pSB1C3_mGMK
Investigator: Chris W., Bea
Vector name: pSB1C3_mGMK upper band 1.1 and pSB1C3_mGMK upper band 1.2 and pSB1C3_mGMK lower band 2.1 and pSB1C3_mGMK lower band 2.2
Mini-Prep following the standart Protokoll
Test digestion:
- Perfomed with 15 µL of total volume with all four clones
| Mastermix/µL | P153/µL | P154/µL | P155/µL | P156/µL | |
| DNA | - | 3,5 | 3,5 | 3,5 | 3,5 |
| BSA (10x) | 7,5 | 4 | 4 | 4 | 4 |
| Buffer 4 (10x) | 7,5 | ||||
| Enzyme XbaI | 2,5 | ||||
| Enzyme AgeI | 2,5 | ||||
| H2O | - | 7,5 | 7,5 | 7,5 | 7,5 |
| Total volume | 15 | 15 | 15 | 15 | 15 |
- All four samples were loaded on a 1% agarose gel
- P153 = pSB1C3_mGMK upper band 1.1
- P154 = pSB1C3_mGMK upper band 1.2
- P155 = pSB1C3_mGMK lower band 1.1
- P156 = pSB1C3_mGMK lower band 1.2
- Plasmid used: P156: pSB1C3_mGMK lower band 1.2
- Primer used: VR-2
- Tube name: Bea_1
cell culture
Investigator: Adrian
Plan for the next virus production procedures
The Motivation: investigation of the influence of different GOI amounts.
The Plan: keep rep/cap and pHelper amount stable (3,3 µg each => 6,6 µg), differ the GOI (YFP) amount in each stock.
Viral Stocks:
- 3,3 µg YFP + 3,3 µg pHelper + 3,3 rep/cap
- 10 µg YFP + 3,3 µg pHelper + 3,3 rep/cap
- 20 µg YFP + 3,3 µg pHelper + 3,3 rep/cap
- 3,3 µg pHelper + 3,3 rep/cap (without GOI !!!)
- one GMK_TK clone (with the confirmed sequence) + 3,3 µg pHelper + 3,3 rep/cap
Transduction plan
| A | 150µl AAV stock 1 | 300µl AAV stock 1 | control no Virus |
|---|---|---|---|
| B | 150µl AAV stock 2 | 300µl AAV stock 2 | 150µl AAV without GOI |
Repetition of cloning of SDM SspI and SDM PvuII
Investigator: Jessica
- Vector: name: pSB1C3_SDM_SspI P125
- Insert: name: pSB1C3_SDM_PvuII P129
- new vector name: pSB1C3_RFC25_SDM_SspI/PvuII P157
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab: 141) BtsI ; Enzyme 2 (no.Lab: 144) BpmI
- DNA concentration (vector): 321,9 ng/µl ; DNA concentration (insert): 308,4 µg/µl
| components | volume of pSB1C3_SDM_SspI /µl | volume of pSB1C3_SDm_PvuII /µl |
| DNA | 4,66 | 4,87 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme ClaI (no.Lab:152) | 1 | 1 |
| Enzyme BpmI (no.Lab:144) | 1 | 1 |
| H2O | 9,34 | 9,13 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
incubation time: 1,5h
0,5 g Agarose,50 ml TBE (1%), 3 µl GELRED (gel was shared with Anna) , at Volt, running time:
Loading plan for agarose gel:
Marker used: GeneRuler ladder mix (Fermentas)
| Marker | Sample 125, 20µl | Sample 129, 20µl | |
|---|---|---|---|
| Lane | 1 | 3 | 5 |
This appproach also didn't work, same result. the guess is that BpmI doesn't work anymore. will be repeated by Chris W. on 05.08.10. the result will show that the guess is right and BpmI from labstock is changed out with a new one.
Test transformation of XL1 competent cells
Investigator: Kira
Test transformation was performed in order to test the efficiency of produced chemical competent XL1 blue cells.
2 plates were prepared: one with 50 pg and the second with 100 pg pUC 18 plasmid (50 pg/㎕) 50 pg plate: 1 ㎕ pUC + 50 ㎕ XL1B cells 100 pg plate: 2 ㎕ pUC + 50 ㎕ XL1B cells
Transformation was performed according to the standard protocol and the plates were incubated at 37 C.
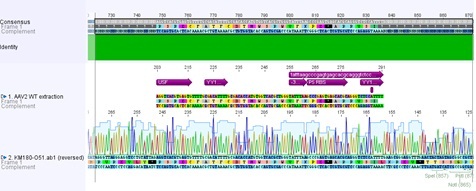
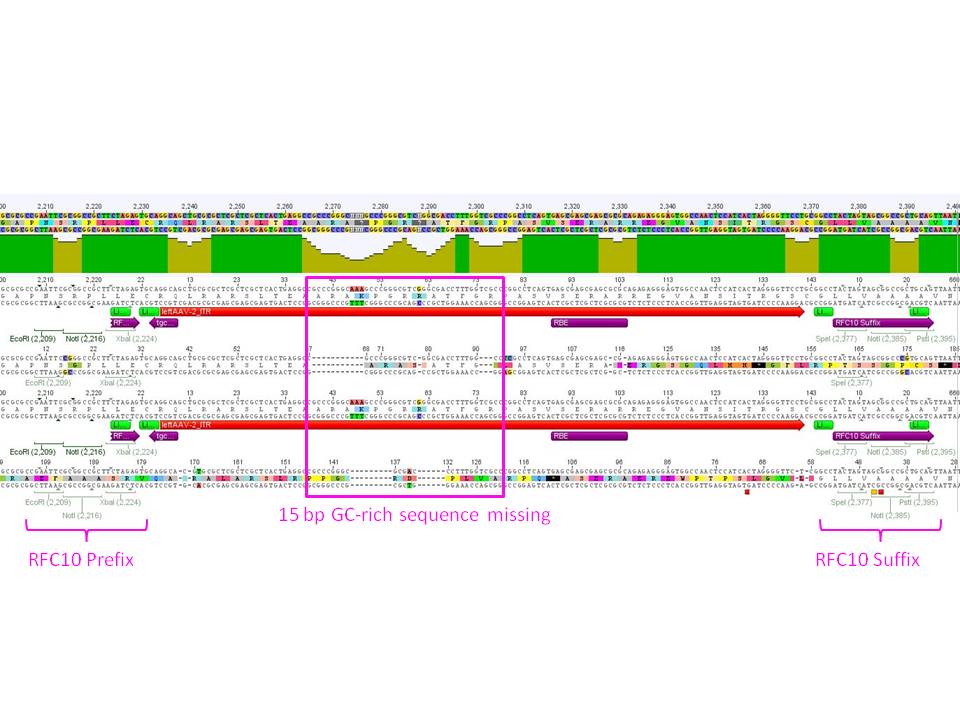
Sequencing results of ITRs
Investigator: Hanna
8 sequencing files were analyzed "per hand" base by base. Alignments (will be inserted soon) delivered that the right ITR is 100% OK and was successfully converted into the RFC10 BioBrick standard.
The sequencing and alignments of the left ITR delivered that a 15 bp fragment in the middle of the sequence is lacking. Therefore further test digestions have to be performed. Nevertheless also this ITR was successfully converted into the RFC10 BioBrick standard. Secondary structure analysis showed that the stemloop-structure will be nevertheless forming. We will try to test whether the referring fragment is also missing in the pAAV_MCS. If it's also lacking there we will continue with this ITR and test whether it functions.
Cloning of Rep68, Rep78 and AAP into pSB1C3
Investigator: Anna
Comments: The PCR of Rep 40/52 didn't work (see 03.08), whereas the PCR of Rep 68/78 and AAP was succesful. Ligation was done with two different samples of pSB1C3 (see agarose gel). Samples from digestion and from ligation are stored in the 4°C freezer.
- Digestion of PCR products and vector:
| components | PCR product /µl | vector /µl |
| DNA | 19 | 10 |
| BSA (10x) | 3 | 3 |
| Buffer 4 (10x) | 3 | 3 |
| Enzyme XbaI | 1 | 1 |
| Enzyme SpeI | 1 | 1 |
| H2O | 3 | 12 |
| Total volume (e.g. 15,20,25,30 µl) | 30 | 30 |
Comments: Digestion was done with XbaI and SpeI, it has to be checked if the inserts are cloned into the vector in the right orientation.
- Purification of Rep68, 78 and AAP:
For the purification 95 µl of buffer PBI was used.
c(Rep68)= 17,6 ng/µl
c(Rep78)= 80,60 ng/µl
c(AAP)= 77,78 ng/µl
- Gelextraction of pSB1C3_RFC25_CFP:
0,5 g Agarose,50 ml TBE (1%), 3 µl GELRED , at 115 Volt, running time:55
5µl loading dye (6x) for the sample, Marker: GeneRuler ladder mix (Fermentas)
c(pSB1C3)= 2,87 ng/µl
c(pSB1C3_2)= 9,46 ng/µl
- Quickligation of PCR products and vector:
For the Ligation 10µl buffer (2x) and 1µl Quickligase were used.
| Sample-no. | vector /µl | insert /µl | |
| pSB1C3 + Rep68 | 1.1 | 6,51 | 2,4 |
| pSB1C3_2 + Rep68 | 1.2 | 3,9 | 5,02 |
| pSB1C32 + Rep78 | 2.1 | 8,2 | 0,8 |
| pSB1C3_2 + Rep78 | 2.2 | 6,82 | 2,18 |
| pSB1C3 + AAP | 3.1 | 8,71 | 0,92 |
| pSB1C3_2 + AAP | 3.2 | 8,1 | 0,9 |
| Total volume (e.g. 15,20,25,30 µl) | 9 |
- Transformation:
The transformation was done following the standard protocol using B21 cells.
80.Labortag 05.08.2010
New LB Agar was prepared.
Investigator: Patrick
Sequenc analysis of BioBricks: CMV, hGH and beta globin
Investigator: Bea
Comments: Sequence analysis of three BioBricks which were cloned into the iGEM standard plasmid pSB1C3. Cloning of this three plasmids were performed at:
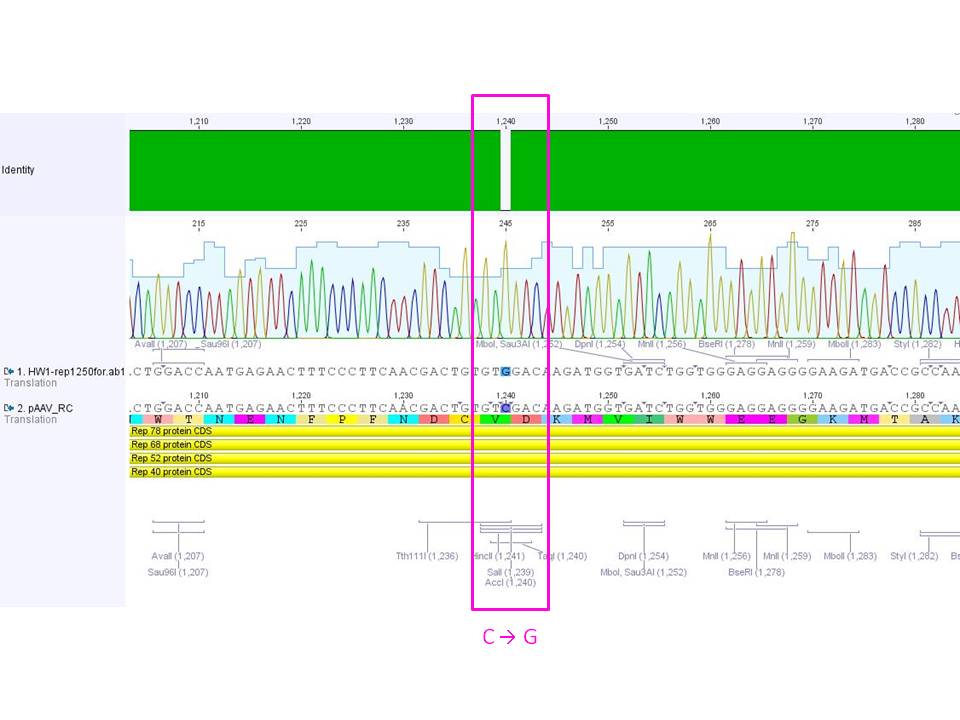
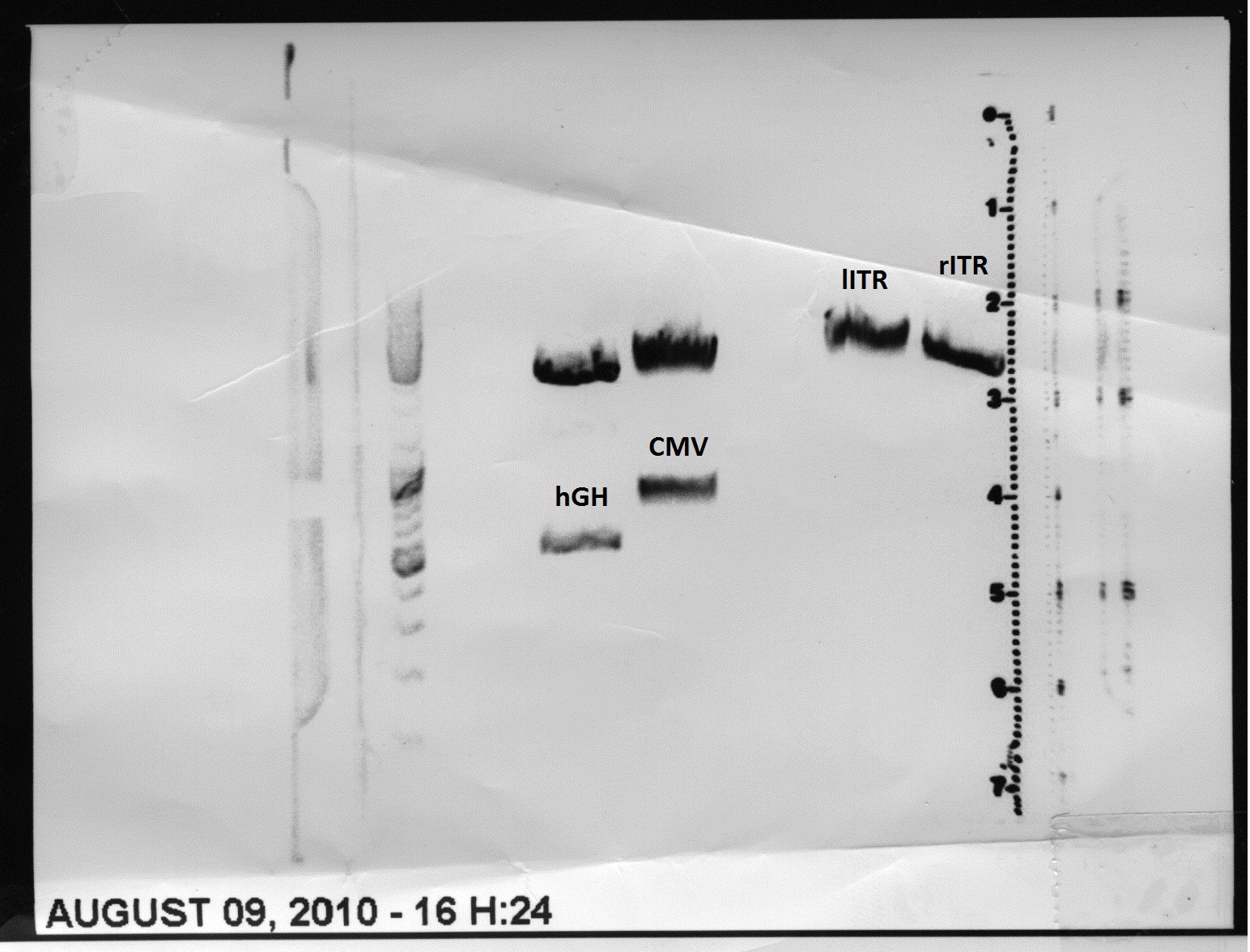
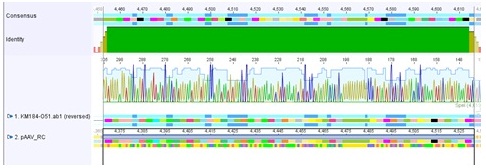
Sequencing results of pSB1C3_CMV:
- Plasmid sent for sequencing:
- Primer used: VR-2
- Results: Sequence read looks good. The incorporation of the PCR product can be confirmed. It can be seen that the insert is in the RFC10 standard and prefix and suffix have the right sequence. Therefore, this BioBrick can be send to the registry and used for BioBrick assembly.But: there is a mutation in the termintor (sequence picture do not show this mutation)
<img src="http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/7/74/Freiburg10_Sequence_analysis_of_PSB1C3_CMV.jpg" />
Sequencing results of pSB1C3_hGH:
- Plasmid sent for sequencing:
- Primer used: VR-2
- Results: Sequence read looks good. The incorporation of the PCR product can be confirmed. It can be seen that the insert is in the RFC10 standard and prefix and suffix have the right sequence. Therefore, this BioBrick can be send to the registry and used for BioBrick assembly.
http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/e/e1/Freiburg10_Sequence_analysis_of_PSB1C3_betaglobin.jpg
Sequencing results of pSB1C3_hGH:
- Plasmid sent for sequencing:
- Primer used: VR-2
- Results: Sequence read looks good. The incorporation of the PCR product can be confirmed. It can be seen that the insert is in the RFC10 standard and prefix and suffix have the right sequence. Therefore, this BioBrick can be send to the registry and used for BioBrick assembly.
http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/4/44/Freiburg10_Sequence_analysis_of_PSB1C3_hGH.jpg
Cellculture
Investigator: Adrian
The Motivation: Transfection
The Action: HEK cells were split into four T75 flasks
The Plan: The cells should be ready at saturday for seeding and monday for transfection
We recived new HT1080 cells, they'll be split tomorrow (thx to Sven)
We recived new tumor cells which overexpress EGFR (thx to barbara)
Picking clones of pSB1C3_AAP_2, pSB1C3_AAP, pSB1C3_Rep78_2, pSB1C3_Rep78, pSB1C3_Rep68_2 and pSB1C3_Rep68
Investigator: Anissa
Of each construct two clones were picked. plates are still stored in the cold-room, tomorrow mini-prep will be done.
PCR and ligation for biobrick-production of pSB1C3_hTERT
Investigator: Anissa,Kerstin
Comments: PCR was performed one time without DMSO and one time with DMSO. Only the approach with DMSO showed bands in the analytic gel after PCR. That's why only this approach will be noted.
- PCR:
(was performed following the standard protocol)
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O111 (phTERT_prefix_for_RFC10) | 2,5 |
| reverse primer: O112 (phTERT_suffix_rev_RFC10) | 2,5 |
| DNA Template | 0,35 µl |
| DMSO (2%) | 1 |
| Phusion Polymerase | 0,5 |
| H2O | 32,15 |
| Total volume | 50 |
PCR program:
| PCR Program | temperature/ °C |
| 1 | 98 |
| 2 | 98 |
| 8x | 58 |
| 3 | 72 |
| 4 | 98 |
| 17x | 70 |
| 5 | 72 |
| 6x | 72 |
| Hold | 4 |
Gel extraction
Gel measurement:
| Sample | Weight | Volume | Concentration |
| hTERT | 0,15g | 20 µl | 37,49 ng/µl |
| pSB1C3_cut | 0,08g | 20 µl | 4,56 ng/µl |
- Digestion of PCR product and vector:
| components | PCR product /µl | vector /µl |
| DNA | 18 | 7,58 |
| BSA (10x) | 0,3 (100X used) | 2 |
| Buffer 4 (10x) | 2,5 | 2 |
| Enzyme XbaI | 1 | 1 |
| Enzyme PstI | 1 | 1 |
| H2O | 2,2 | 6,42 |
| Total volume (e.g. 15,20,25,30 µl) | 25 | 20 |
Ligation with T4-Ligase and transformation with BL 21 was made. Clones have to be picked tomorrow.
Transformation evaluation of XL1B cells and repetition of transformation
Investigator: Kira
Against our expectations both agar plates contain very few colonies. In order to figure out if the lack of colonies is due to the XL1 blue cells or loss of pUC activity, test transformation will be repeated this evening with recently prepared XL1 blue cells as well as with Lab-XL1B cells.
50 pg pUC-plate contains 8 colonies
100 pg pUC-plate contains 18 colonies
Transformation will be performed again with 100 pg pUC (= 2 ㎕ pUC).
Cloning of SDM SspI and SDM PvuII
Investigator: Chris W.
- Vector: name: pSB1C3_SDM_SspI P125
- Insert: name: pSB1C3_SDM_PvuII P129
- new vector name: pSB1c3_SDM_SspI/PvuII P157
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab: 144) BpmI ; Enzyme 2 (no.Lab: 152) ClaI
- DNA concentration (vector): 321,9 ng/µl ; DNA concentration (insert): 308,4 µg/µl
| components | volume of pSB1C3_SDM_SspI /µl | volume of pSB1C3_SDm_PvuII /µl |
| DNA | 4,66 | 4,87 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme BpmI (no.Lab:144) | 1 | 1 |
| Enzyme ClaI (no.Lab:152) | 1 | 1 |
| H2O | 9,34 | 9,13 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Comment: I guess enzyme 144 was BpmI and enzyme 152 was ClaI ? (Jessica)
Comment: u r right, changed (Chris)
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 130 Volt, running time:45
Loading plan for agarose gel:
Marker used: GeneRuler ladder mix (Fermentas)
| Marker | Sample 125, 20µl | Sample 129, 20µl | |
|---|---|---|---|
| Lane | 1 | 5 | 7 |
The mutual vector (now without SspI) and insert (now without PvuII) were cut out. The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P125 Sspl: 12,8 ng/µl
- P129 PvuII: 11,6 ng/µl
The Ligation was performed as following:
- Vector Volume: 2,81 µl
- Insert Volume: 5,19 µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubate for 30min
Transformation: performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol
Miniprep of p158 and p159
Plasmid Mini-Prep
Investigator: Bea & Volker
For the tripple mutant of pAAV-RC a Quick-change reaction was carried out to remove the last restriction site (SalI) that is required for the Viral Brick standard. For this construct glycerol stocks and minipreps were carried out for two clones.
Glycerol Stocks
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| Plasmidname | pAAV_RC_1.2 SDM SalI | align="left"| pAAV_RC_1.2 SDM SalI |
| Date | 05.08.2010 | 05.08.2010 |
| given glycerol-stock no. | B126 | B127 |
| given plasmid no. | p158 | p159 |
TO do: Test digestion and sequencing!!!
81.Labortag 06.08.2010
Cloning Rep40 & Rep52 into pSB1C3
Intention: get biobrick ready
Investigator: Patrick
PCR of pKEX-2XL.Rep 40 (P22) and pKEX-2XL.Rep 52 (P23) following gelrun (1%) and gelextraction.
Used Primers:
- Praefix Rep40_52ex (O94)
- Suffix Rep 40_68ex (O96)
- Suffix Rep 52_78ex (O97)
PCR programm of Rep40:
- 98°C 1 min
- 98°C 15 sec
- 63°C 25 sec
- 72°C 15 sec Repeat this cycle 8 times.
- 98°C 15 sec
- 68°C 25 sec
- 72°C 15 sec Repeat this cycle 17 times.
- 72°C 5 min
- Hold 4°C
PCR programm of Rep52:
- 98°C 1 min
- 98°C 15 sec
- 62°C 25 sec
- 72°C 20 sec Repeat this cycle 8 times.
- 98°C 15 sec
- 66°C 25 sec
- 72°C 20 sec Repeat this cycle 17 times.
- 72°C 5 min
- Hold 4°C
| ingredients | Rep 40 | Rep40 + DMSO | Rep52 | Rep52 + DMSO |
| 5x Phusion HF buffer | 10 µl | 10 µl | 10 µl | 10 µl |
| 10 mM dNTP mix | 1 µl | 1 µl | 1 µl | 1 µl |
| for Primer O94 (1:10 dilution, 05 µM) | 2,5 µl | 2,5 µl | 2,5 µl | 2,5 µl |
| rev Primer (1:10 dilution, 0,5 µM) | 2,5 µl O96 | 2,5 µl O96 | 2,5 µl O97 | 2,5 µl O97 |
| DNA template | 1 µl, 112,7 ng/µl | 1 µl, 112,7 ng/µl | 0,9 µl, 142,3 ng/µl | 0,9 µl, 142,3 ng/µl |
| DMSO | 0 µl | 0,5 µl | 0 µl | 0,5 µl |
| Phusion Polymerase | 0,5 µl | 0,5 µl | 0,5 µl | 0,5 µl |
| H2O | 32,5 µl | 32 µl | 32,6 µl | 32,1 µl |
| Total volume | 50 µl | 50 µl | 50 µl | 50 µl |
Ecpected size of Rep40: 940 bp, expected size of Rep52:1198 bp. There are two samples of Rep40 and Rep52 and the PCR of one of each was run with 1% DMSO because the Rep40 Praefix has a strong secondary structure and was used as a primer for Rep 40 and Rep 52

Digestion of pSB1C3 (P51.1) with EcoRI-HF and SpeI following gelrun (0.8%) and gelextraction.
Expected size of the fragments: about 2000 bp and 800bp

All marked fragments were extracted. The Gelextraction was performed according to the standard protocol following a digestion of the PCR products with EcoRI HF and SpeI and a purification of the PCR product. The Ligation was not performed according to the standard protocol: 1 µl T4 DNA Ligase, 1 µl (10x) Buffer, 3 µl vector pSB1C3 (60 ng/µl) and 5 µl insert (all concentrations about 100 ng/µl). The Transformation (with BL21) was performed according to the standard protocol. Tomorrow there will hopefully be some clones on the agar plates.
BioBrick Assembly of pSB1C3_beta globin_mVenus
Investigator: Bea
Comments: BioBricks are ready to use. The sequenced pSB1C3_beta globin (P133) and pGA14_mVenus (P60) will be digested with different enzymes and will be assembled in order to produce the first step in assembling the whole vector together.
- First the plasmids P133 and P60 were digested:
| pSB1C3_betaglobin/µL | pGA14_mVenus/µL | |
| DNA | 7 | 9 |
| BSA (100x) | 0,2 | 0,2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme XbaI | 1 | 1 |
| Enzyme AgeI | 1 | 1 |
| H2O | 8,8 | 6,8 |
| Total volume | 20 | 20 |
- Incubation of plasmids at 37°C for 2 hours
- Load samples on preparative 1% agarose gel and run at 110 V, 45 minutes
Results: As it can be seen in the picture below, digestion of the vector pSB1C3_betaglobin (left lane) revealed a band at around 2500 - 3000 bp. The expected size after digetsing with SpeI and PstI was: 2555 bp. The band ran a little bit higher than expected, anyhow the band was cut out of the gel. The right lane belongs to the pGA14_mVenus which was digested with XbaI and PstI which resulted in two fragments. mVENUS was digested. Expected sizes were: mVenus = 756bp and pGA14 = 2900bp. The expected sizes correspond to the bands seen in the picture below. The smaller fragment belonging to mVenus was cut out of the gel as well.

After gel extraction has been performed, the concentrations of the two fragments were measured and we obtained following concentrations:
- c(pSB1C3_betaglobin) = 15,62 ng/µL
- c(mVenus) = 6,83 ng/µL
For ligation the T4-Ligase were used.
- v(pSB1C3_betaglobin) = 2,64 µL
- v(mVenus) =5,36 µL
- v(10xT4 Ligase buffer) = 1 µL
- v(T4-Ligase) = 1 µL
- Total volume = 10 µL
Additionally a control ligation was performed.
- v(pSB1C3_betaglobin) = 2,64 µL
- v(H20) =5,36 µL
- v(10xT4 Ligase buffer) = 1 µL
- v(T4-Ligase) = 1 µL
- Total volume = 10 µL
- Ligation duration was 45 minutes on room temperature.
After ligation a transformation was performed.
- Used bacterial strain: XL1-B
- LB-agar plates containing chloramphenicol were plated and put in the 37°C room over night
To do: Mini-Prep and test digestion in order to verify assembly of beta globin and YFP. After verification of the fusion of the two fragments this construct is ready for the next BioBrick assembly step.
Test digestion of SalI SDM
Investigator: Hanna
Test digestion
- buffer used: 3 ; Restriction-enzymes used: Enzyme 1 (no. Lab: 53): SalI ; Enzyme 2: XbaI
- Plasmid
- Given Plasmid-Number: P158; DNA concentration: 398.8 ng/µL ;
- Given Plasmid-Number: P159; DNA concentration: 437.4 ng/µL ;
| Components | P158 Volume/µL | P159 Volume/µL |
| DNA | 2.5 | 2.3 |
| BSA (10x) | 1 | 1 |
| Buffer 3 (10x) | 1 | 1 |
| SalI (no. Lab: 53) | 0.5 | 0.5 |
| XbaI | 0.75 | 0.75 |
| H2O | 4.25 | 4.45 |
| Total volume | 10 | 10 |
Incubation: 1 h
Agarose-Gel:
0.5 g Agarose, 50 mL TBE (1 %),3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (if SDM didn't work) | Expected size 2 (if SDM didn't work) | Expected size 3 (if SDM didn't work) |
|---|---|---|---|---|---|
| P158 | 10 µl | 2 µl | 6129 bp | 1143 bp | 61 bp |
| P159 | 10 µl | 2 µl | 6129 bp | 1143 bp | 61 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P158 /µl | Sample P159 /µl | Marker /µL | |
|---|---|---|---|---|
| Lane | 6.5 | 10 | 10 | 7 |
Comments: Test digestion looked good: No 1143 bp fragment was detectable in neither test digestion.
P158 was sent for sequencing to GATC.
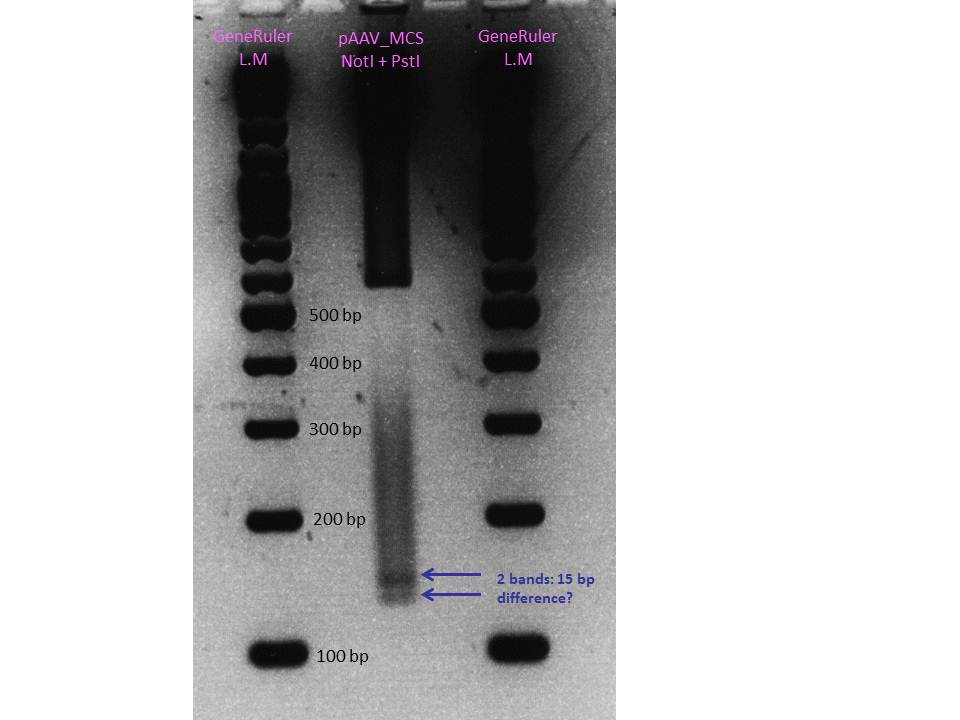
ITR test digestion of pAAV_MCS
Investigator: Hanna
Comment: Because sequencing of the right ITR delivered that perhaps a 15 bp fragment is missing in the sequence, we wanted to check, whether this fragment is also not present in the original pAAV_MCS plasmid or whether the loss of these 15 bp happened during cloning.
Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1: NotI; Enzyme 2: PstI
- Plasmid
- Plasmid-Number: from Sven; DNA concentration: 259 ng/µL ;
| Components | Volume/µL |
| DNA | 3.8 |
| BSA (10x) | 1.5 |
| Buffer 4 (10x) | 1.5 |
| NotI-HF | 0.75 |
| PstI-HF | 0.75 |
| H2O | 3.2 |
| Total volume | 15 |
Incubation: 1 h
Agarose-Gel:
0.5 g Agarose, 50 mL TBE (1 %),3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size ITRs | Expected size 2 | Expected size 3 | Expected size 4 |
|---|---|---|---|---|---|---|
| pAAV_MCS | 15 µl | 2 µl | 145 bp | 1216 bp | 555 bp | 2608 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample /µl | Marker /µL | |
|---|---|---|---|
| Lane | 6.5 | 12 | 6.5 |
Comments: The test digestion showed that there're two bands between the 100 and 200 bp marker nucleotides! By comparing the gel picture with the last mini-prep digestion of the fancy method we found out that the space between the two bands are similar. Because of that we assumed that the 15 pb are already missing in the original Stratagene plasmid!!!
Conclusion: Because we already tested the Stratagene Kit via YFP expression, we can conclude that the usage of the "wrong" rigth ITR works. Therefore we decided to continue working with this "mutated" = ENGINEERED :) right ITR as BioBrick.
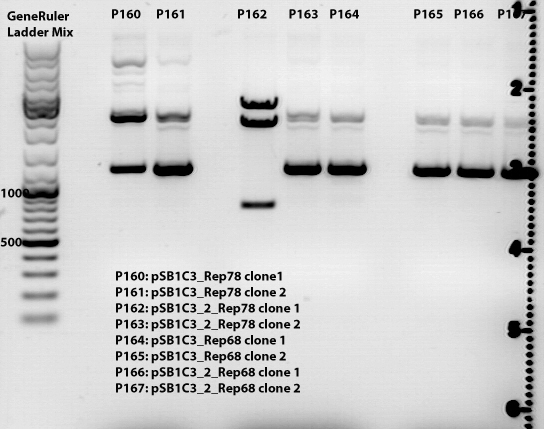
Miniprep of pSBC13_Rep78, pSB1C3_Rep68, pSB1C3_AAP, pSB1C3_rightITR and psB1C3_leftITR
Plasmid Mini-Prep
Investigator: Kerstin
Glycerol Stocks
pSB1C3_Rep78
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B128 | B129 |
| given plasmid no. | P160 | p161 |
| DNA-concentration ng/µl | 139,1 | 71,0 |
pSB1C3_2_Rep78
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B130 | B131 |
| given plasmid no. | P162 | p163 |
| DNA-concentration ng/µl | 112,1 | 79,7 |
pSB1C3_Rep68
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B132 | B133 |
| given plasmid no. | P164 | p165 |
| DNA-concentration ng/µl | 80,2 | 72,8 |
pSB1C3_2_Rep68
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B134 | B135 |
| given plasmid no. | P166 | p167 |
| DNA-concentration ng/µl | 85,6 | 69,1 |
pSB1C3_AAP
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B136 | B137 |
| given plasmid no. | P168 | p169 |
| DNA-concentration ng/µl | 63,6 | 56,3 |
pSB1C3_2_AAP
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B138 | B139 |
| given plasmid no. | P170 | p171 |
| DNA-concentration ng/µl | 135,1 | 114,2 |
pSB1C3_rightITR
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B140 | B141 |
| given plasmid no. | P172 | p173 |
| DNA-concentration ng/µl | 73,7 | 66,4 |
pSB1C3_leftITR
| Clone 1 | Clone 2 | |
| Bacteria strain | BL-21 | BL-21 |
| given glycerol-stock no. | B142 | B143 |
| given plasmid no. | P174 | p175 |
| DNA-concentration ng/µl | 74,9 | 80,3 |
Test digestion:
pSB1C3_Rep78, pSB1C3_Rep68 and pSB1C3_AAP were digested with XbaI and SpeI
pSB1C3_rightITR and pSB1C3_leftITR were digested with EcoRI and PstI.
| components | volume of P160/µl | volume of P161/µl | volume of P162/µl | volume of P163/µl | volume of P164/µl | volume of P165/µl | volume of P166µl | volume of P167/µl | volume of P168µl | volume of P169/µl | volume of P170/µl | volume of P171/µl | volume of P172/µl | volume of P173µl | volume of P174/µl | volume of P175/µl |
| DNA | 5,8 | 11,3 | 7,1 | 10 | 9,9 | 10,9 | 9,3 | 11,6 | 12,5 | 14,2 | 5,9 | 7,0 | 10,9 | 12,0 | 10,7 | 9,7 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| Enzyme 2 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 9,2 | 3,7 | 7,9 | 5,0 | 5,1 | 4,1 | 5,7 | 3,4 | 2,5 | 0,8 | 9,1 | 8 | 4,1 | 3 | 4,3 | 5,3 |
| Total volume /µl | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
Expected sizes:
- Rep78: 1,8 kb
- Rep68: 1,6kb
- AAP: 650bp
- left ITR: 150bp
- right ITR: 150bp
Results of samples loaded 1,5% agarose gel and ran ~50 minutes at 115 V:
Results of samples loaded 0,8% agarose gel and ran ~50 minutes at 115 V:
Sequencing
P160, P165 and P170 were send for sequencing.
82.Labortag 07.08.2010
Cellculture
HEK 293 cells were split and seeded according to the standard protocol: 2x T75 flask and 10x10cm cellculture dishes. Investigator: Patrick
Continuation: Cloning Rep40 & Rep52 into pSB1C3
Investigator: Patrick
Results: no clones could be picked because there were no clones. Maybe the PCR purification or one of the following steps did not work (the PCR purifications were quite bad) so i purified the PCR products again yielding very low DNA concentrations (0-13,28 ng/µl) and still very obvious pollution of the sample. Nonetheless a ligation with the insert Rep40+DMSO, Rep52, Rep52+DMSO and the vector pSB1C3 was carried out following a transformation and according to the standard protocols. Hopefully there will be some clones tomorrow.
Mini-Prep and test digestion of pSB1C3_SDM_SspI/PvuII
Investigator: Jessica
Plasmid Mini-Prep
- new vector name: pSB1C3_SDM_SspI/PvuII
Glycerol Stocks
pSB1C3_SDM_SspI/PvuII
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | BL21 | BL21 | BL21 |
| Plasmidname | pSb1C3_SDM_SspI/PvuII | pSb1C3_SDM_SspI/PvuII | pSb1C3_SDM_SspI/PvuII |
| Date | 7.8.10 | 7.8.10 | 7.8.10 |
| given glycerol-stock no. | B144 | B145 | B146 |
| given plasmid no. | P157.1 | P157.2 | P157.3 |
Test digestion
- buffer used: 2 ; Restriction-enzymes used: Enzyme 1 SspI ; Enzyme 2 PvuII
- Plasmid: pSB1C3_SDM_SspI/PvuII:
- Given Plasmid-Number: P157.1; DNA concentration: 246,3 ng/µL ;
- Given Plasmid-Number: P157.2; DNA concentration: 231,7 ng/µL ;
- Given Plasmid-Number: P157.3; DNA concentration: 219,6 ng/µL ;
| Components | P157.1 Volume/µL | P157.2 Volume/µL | P157.3 Volume/µL |
| DNA | 2,8 | 3,0 | 3,19 |
| BSA (10x) | - | - | - |
| Buffer no. 2 (10x) | 1 | 1 | 1 |
| Enzyme 1 SspI | 0.5 | 0.5 | 0.5 |
| Enzyme 2 PvuII | 0.5 | 0.5 | 0.5 |
| H2O | 5,2 | 5 | 4,81 |
| Total volume | 10 | 10 | 10 |
- Incubation: 45 min
Agarose-Gel:
0.45 g Agarose, 50 mL TAE, 3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl |
|---|---|---|
| P157.1 | 10 µl | 1,6 µl |
| P157.2 | 10 µl | 1,6 µl |
| P157.3 | 10 µl | 1,6 µl |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P157.1 /µL | Sample P157.2 /µL | Sample P157.3 /µL | |
|---|---|---|---|---|
| Lane | 7 | 10 | 10 | 10 |
I expect one cut from the PvuII RS in the CFP --> linearized
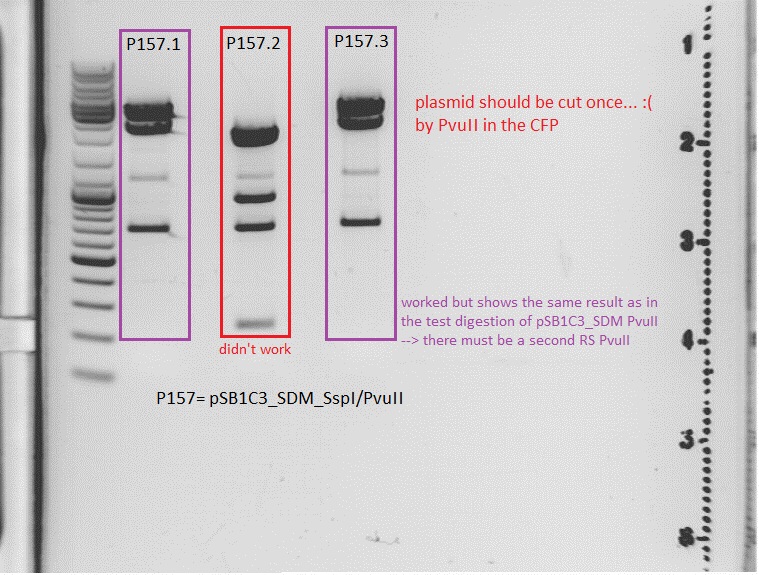
Comment:
Comments:Oh f... it makes no sense... if the cloning didn't work, there can just be 3 bands (one time SspI and twice PvuII). The results of the test digestion from aug 1th showed that there can be one RS PvuII more. but then you have a maximum of 4 bands. P157.2 show 5 bands. i can't interpret this result at the moment. interpretation will follow
Evaluation of test transformation with XL1 blue cells and pUCI
Investigator: Kira
Transformation was performed with lab XL1B and iGEM XL1B cells. iGEM plate contained just 25 colonies, while lab plate contained around 50 colonies. Conclusion: the production of competent cells has to be repeated.
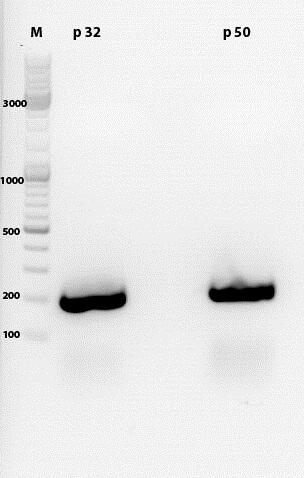
Biobrick production of p5 promoter from pTAV2 and pAAV_RC
Investigator: Kira
- PCR:
DNA samples were diluted 1:100
| Ingredients | p 32 | p 50 |
| 5X Phusion HF buffer | 10 µl | 10 µl |
| 10 mM dNTP mix | 1µl | 1 µl |
| forward primer: O114 | 2,5µl | 2,5 µl |
| reverse primer: O115 | 2,5 µl | 2,5 µl |
| DNA Template | 0,3 µl | 0,4 µl |
| DMSO | 0 µl | 0 µl |
| Phusion Polymerase | 0,5 µl | 0,5 µl |
| H2O | 33,2 µl | 28,6 µl |
| Total volume | 50 µl | 50 µl |
PCR program:
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 30" |
| 60°C | 25" | |
| 72°C | 6" | |
| 17x | 98°C | 30" |
| 70°C | 25" | |
| 72°C | 6" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Digestion of plasmid backbone:
c (pSB1C3) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA 1 µg | 6,0 µl |
| BSA (100x) | 0,2 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 XbaI | 1 µl |
| Enzyme 2 PstI HF | 1 µl |
| H2O | 9,8 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
1% agarose gel with PCR products:
Concentrations after gel extraction:
c(p32) = 67,95 ng/µl
c(p50) = 32,45 ng/µl
c(vector) = 7,54 ng/µl
Digestion of PCR products:
| Components | PCR product Volume/µL |
| DNA 1 µg | 23,0 µl |
| BSA (100x) | 0,3 µl |
| Buffer no. 4 (10x) | 3,0 µl |
| Enzyme 1 XbaI | 1,5 µl |
| Enzyme 2 PstI HF | 1,5 µl |
| H2O | 0,7 µl |
| Total volume | 30 |
Concentrations after PCR purification:
c(p32) = 21, 63 ng/µl
c(p50) = 19,48 ng/µl
Ligation:
Quickligase was used for ligation.
p32 DNA-Mix: 8 µl vector + 1 µl insert
p50 DNA-Mix: 7,9 µl vector + 1,1 µl insert
Transformation:
Transformation was performed according to the standard protocol with DYT and BL21 cells. The cells were spread on the agar plates containing chloramphenicol.
Sequencing results of SalI SDM
Investigator: Hanna
Comment: The test digestion already showed that the SDM of the SalI restriction site in pAAV_RC worked. This could be validated by sequencing the plasmid with the Rep-1250_for primer:
P58 can be used for further RepCap-experiments.
83. Labortag 08.08.2010
HT 1080 cells were split and seeded: 3x10^6 cells per into each well of a 6-well plate.
A431 cells were washed and received new medium.
There were no clones on the agar plates so they were put back into the 37°C room. Maybe there will be some clones tomorrow.
84. Labortag 09.08.2010
Midi-Prep Inoculation
- pAAV_RC
- pAAV_RC_1.2 SDM SalI (P158)
- pAAV_RC_mGMK_TK30 clone 1 (P81)
- pAAV_RC_mGMK_TK30 clone 2 (P82)
Investigators: Chris W., Patrick
Continuation: Cloning Rep40 & Rep52 into pSB1C3
Investigator: Patrick
Two Rep52+DMSO clones grew. They were picked and tomorrow there will be a midi prep and a test digestion.
Simultaneously a new approach for cloning Rep40 and Rep52 into pSB1C3 was performed. (see next header)
Repetition: BioBrick production of Rep40 & Rep52
Investigator: Anna
Comments: Another approach was done because there were no clones for Rep 40 and only two for Rep 52 (Cloning see 06.08.2010). PCR was performed again with DMSO. Samples from gel extraction are stored in 4°C freezer.
PCR
| Ingredients | Rep 40 | Rep52 |
| 5X Phusion HF buffer | 10 µl | 10 µl |
| 10 mM dNTP mix | 1µl | 1 µl |
| forward primer: O094 | 2,5µl | 2,5 µl |
| reverse primer: O0096/ O097 | 2,5 µl | 2,5 µl |
| DNA Template (1ng) | 1 µl | 1 µl |
| DMSO | 1 µl | 1 µl |
| Phusion Polymerase | 0,5 µl | 0,5 µl |
| H2O | 31,5 µl | 31,5 µl |
| Total volume | 50 µl | 50 µl |
PCR programm
| PCR Program | temperature/ °C | Time | PCR products |
| 1 | 98 | 1min | |
| 2 | 98 | 15s | |
| 8x | *** | 15s | 63°C |
| 3 | 72 | *** | 15s |
| 4 | 98 | 15s | |
| 19x | *** | 15s | 68°C |
| 5 | 72 | *** | 15s |
| 6x | 72 | 5min | |
| Hold | 4 |
Agarose gel
0.5 g Agarose, 50 mL TAE (1 %),3 µL GELRED, at 120 Volt, running time: 45 minutes
| Sample | Sample / µl] | Loading dye (6x) / µl | Expected size |
|---|---|---|---|
| p22 (Rep40) | 44 µl | 7 µl | 940 bp |
| p23 (Rep52) | 44 µl | 7 µl | ~1200 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample Rep40 /µl | Sample Rep52 /µl | |
|---|---|---|---|
| Lane | 7 | 51 | 51 |
Gel extraction
PCR products were eluted with 20 µl BE puffer following the standard protocol.
c(Rep40)= 28,03 ng/µl
c(Rep52)= 121,16 ng/µl
Comments: Digestion was done with the wrong enzymes (EcoRI HF and SpeI), it will be repeated on 11.08. with XbaI and SpeI
Sequence analysis of pSB1C3_mGMK
Investigator: Bea
Comments: mGMK was cloned into the pSB1C3 backbone in order to send it to the parts registry!. The sequence analysis revealed that the mGMK was successfully cloned into pSB1C3 plasmid. This polasmid is in the RFC25 standard.
-
Results sequencing: Sequence read ok!
Primer used: VR-2
Comment: A mutation has been found in the pSB1C3 backone which was found generally in the backbone which we received from the iGEM HQ.
Mini-Prep of pSB1C3_hTERT, pSB1C3_beta-globin_mVenus, pSB1C3_pTAV2(P5), pSB1C3_pAAV_RC(P5TATAles)
Investigator: Jessica
Bold text
Mini-Prep following the standart Protokoll
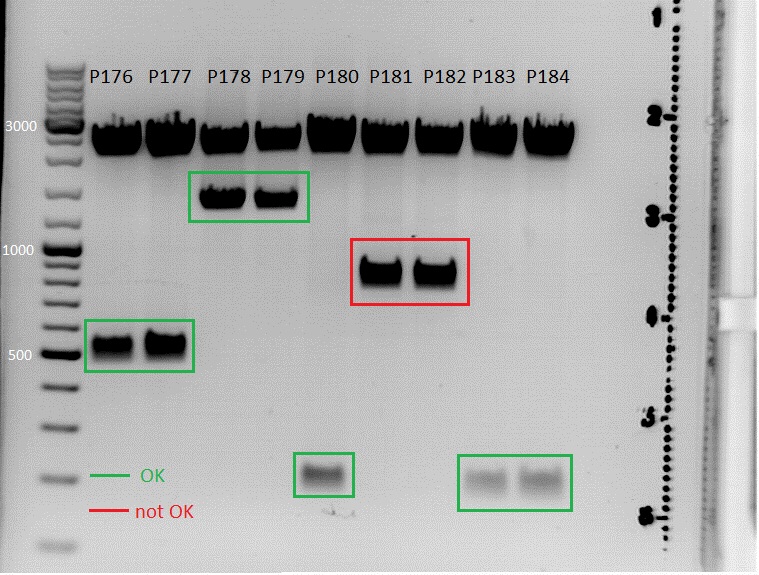
Test digestion:
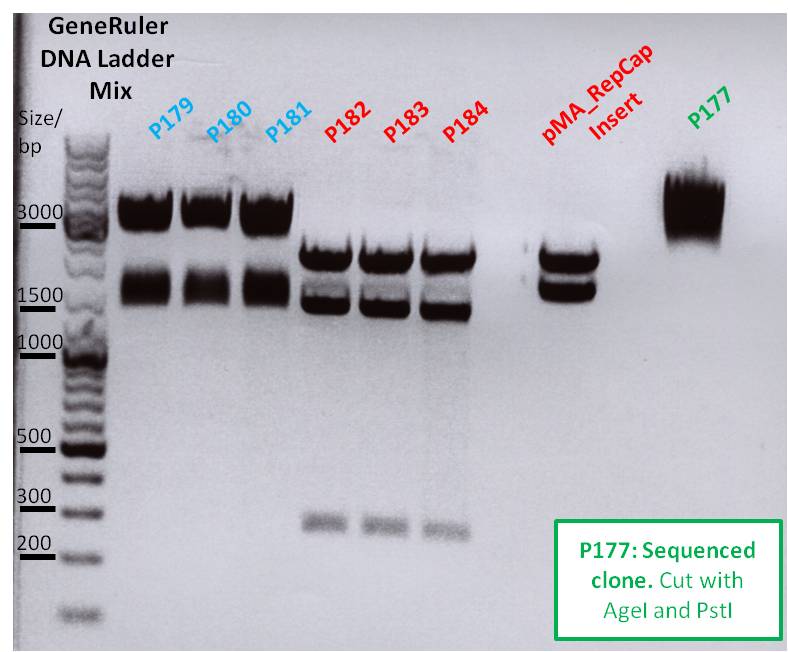
- Perfomed with 10 µL of total volume with all 9 clones
| Mastermix/µL | P176/µL | P177/µL | P178/µL | P179/µL | P180/µL | P181/µL | P182/µL | P183/µL | P184/µL | |
| DNA | - | 4,3 | 2,3 | 3,0 | 2,4 | 6,4 | 5,0 | 4,2 | 4,0 | 4,0 |
| BSA (10x) | 10 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Buffer 4 (10x) | 10 | |||||||||
| Enzyme XbaI | 5 | |||||||||
| Enzyme AgeI | 5 | |||||||||
| H2O | - | 2,7 | 4,7 | 4,0 | 4,6 | 0,6 | 2,0 | 2,8 | 3,0 | 3,0 |
| Total volume | 30 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
- All 9 samples were loaded on a 1% agarose gel
- P176 = pSB1C3_hTERT clone1
- P177 = pSB1C3_hTERT clone2
- P178 = pSB1C3_betaglobin clone1
- P179 = pSB1C3_betaglobin clone2
- P180 = pSB1C3_pTAV2(P5) clone1
- P181 = pSB1C3_pTAV2(P5) clone2
- P182 = pSB1C3_pAAV_RC(P5TATAless) clone1
- P183 = pSB1C3_pAAV_RC(P5TATAless) clone2
- P184 = pSB1C3_pAAV_RC(P5TATAless) clone3
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) |
|---|---|---|---|
| P176 | 10 µl | 1,6 µl | 474 bp |
| P177 | 10 µl | 1,6 µl | 474 bp |
| P178 | 10 µl | 1,6 µl | 509 bp |
| P179 | 10 µl | 1,6 µl | 509 bp |
| P1780 | 10 µl | 1,6 µl | 164 bp |
| P181 | 10 µl | 1,6 µl | 164 bp |
| P182 | 10 µl | 1,6 µl | 164 bp |
| P183 | 10 µl | 1,6 µl | 164 bp |
| P184 | 10 µl | 1,6 µl | 164 bp |
Comments: hTERT and betaglobin_mVenus is inserted in pSB1C3, pTAV(P5) and pAAV_RC (P5TATAless) will be sequenced
Primer: Reverse Primer VR2
Cell culture
Investigators: Adrian, Kerstin, Patrick
Transfection on HEK293
The motivation: first we want to create new stock of AAV without GOI, the next viral stock is with correct TK/GMK, the last stocks are with different YFP-amounts to check if the amount of GOI is a critical step for transduction effency (transgene expression).
| Plate | pAAV_RC/µg | pHelper/µg | GOI/µg |
| 1 | 3,3 | 3,3 | no GOI |
| 2 | 6,6 | 3,3 | no GOI |
| 3 | 3,3 | 3,3 | 3,3 TK_GMK clone1 |
| 4 | 3,3 | 3,3 | 3,3 TK_GMK clone1 |
| 5 | 3,3 | 3,3 | 3,3 TK_GMK clone2 |
| 6 | 3,3 | 3,3 | 3,3 YFP |
| 7 | 3,3 | 3,3 | 10 YFP |
| 8 | 3,3 | 3,3 | 20 YFP |
| 9 | 3,3 | 3,3 | 40 YFP |
| 10 | 3,3 | 3,3 | 60 YFP |
Transduction of HT1080
Motivation
The Motivation: checking survivalrate of Cells, with either no Virus, virus without GOI and virus with YFP (10µg stock)
The Plan:
Plate I
| control, no cells | no GOI 150µl | 10µg 500µl| |
| control, no virus | no GOI 300µl | 10µg 750µl| |
Plate II
| control, no cells | no GOI 150µl | 10µg 500µl| |
| control, no virus | no GOI 300µl | 10µg 750µl| |
Plate III
| control, no cells | no GOI 150µl | 10µg 500µl| |
| control, no virus | no virus | 10µg 750µl| |
Passaging and seeding of A431
one 6er dish was prepared for making pictures and one T75 Flask.
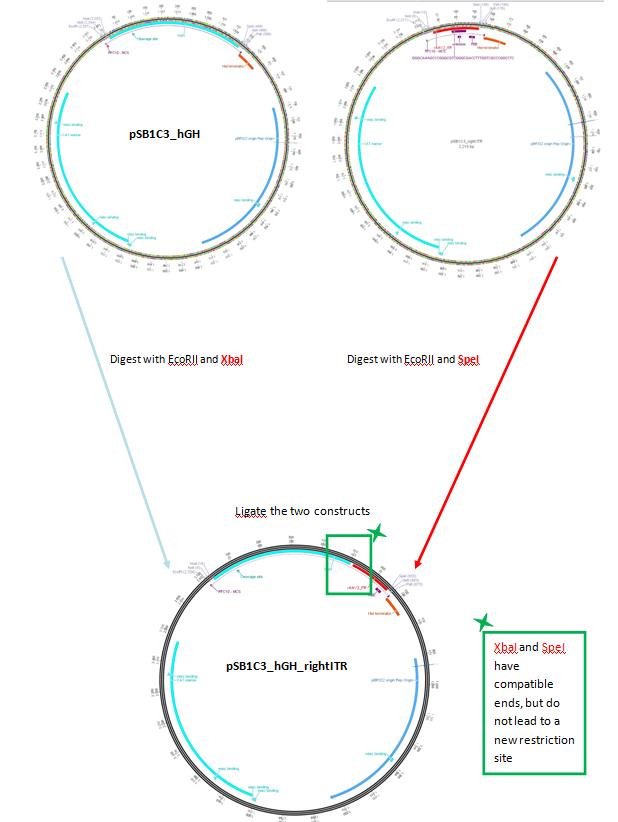
BioBrick Assembly of pSB1C3_leftITR_CMV and pSB1C3_hGH_rightITR
Investigator: Bea, Achim, Jo
Comments: In order to proceed with the BioBrick assembly the pasmids pSB1C3_left ITR and pSB1C3_CMV were used and pSB1C3_rigthITR and pSB1C3_hGH were used in order to obatin plasmids with both sequences.
Digestion:
| components | pSB1C3_left ITR | pSB1C3_CMV | pSB1C3_right ITR | pSB1C3_hGH |
| DNA | 13,4 | 9,5 | 13,6 | 6,6 |
| BSA (10x) | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 |
| Enzyme1 | 1 (SpeI) | 1 (Xba1) | 1 (EcoRI) | 1 (EcoRI) |
| Enzyme2 | 1 (PstI) | 1 (PstI) | 1 (XbaI) | 1 (SpeI) |
| H2O | 0,6 | 4,5 | 0,4 | 7,4 |
| Total volume | 20 | 20 | 20 | 20 |
Preparation of gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 45 minutes
Oh, nice picture.. I can recognize many details :)*g* (Hanna)
Expected sizes of constructs:
- pSB1C3_CMV: 650 bp
- pSB1C3_lITR: 2200 bp
- pSB1C3_hGH: 500 bp
- pSB1C3_rITR: 2200 bp
The correspnding bands were cut out and Gel-Extraction was performed according to protocol.
concentrations measured via NanoDrop:
- pSB1C3_CMV: 13,51 ng/µl
- pSB1C3_lITR: 18,49 ng/µl
- pSB1C3_hGH: 7,22 ng/µl
- pSB1C3_rITR: 20,24 ng/µl
Ligation:
For ligation, T4 ligase was used.
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Ligation was carried out as following:
- Ligation 1:
- pSB1C3_CMV: 4,4 ng/µl
- pSB1C3_lITR: 3,6 ng/µl
- Ligation 2:
- pSB1C3_hGH: 5,25 µl
- pSB1C3_rITR: 2,75 µl
Incubation for 40 minutes.
Transformation:
Transformation carried out according to standard protocol (BL21). Cells were plated on an agar plate with chloramphenicol.
Repetition: Cloning of SDM SspI and SDM PvuII
Investigator: Stefan
Comments: Last week's cloning of SDM_SspI and SDM_PvuII didn't work out, so this is the repetition. In this approach the same restriction enzymes and amounts of DNA were used.
- Vector: name: pSB1C3_SDM_SspI P125
- Insert: name: pSB1C3_SDM_PvuII P129
- buffer used: 4
- Restriction-enzymes used:
- BpmI (no. Lab: 144)
- ClaI (no.Lab: 152)
- DNA concentration (P125): 321,9 ng/µl
- DNA concentration (P129): 308,4 ng/µl
Digestion:
| components | volume of pSB1C3_SDM_SspI /µl | volume of pSB1C3_SDm_PvuII /µl |
| DNA | 4,66 | 4,87 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| BpmI (no.Lab:144) | 1 | 1 |
| ClaI (no.Lab:152) | 1 | 1 |
| H2O | 9,34 | 9,13 |
| Total volume | 20 | 20 |
Gel extraction:
Preparation of gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 45 minutes
Expected sizes of constructs:
- pSB1C3_SDM_SspI: 1819 bp
- pSB1C3_SDM_PvuII: 981 bp
The correspnding bands were cut out and Gel-Extraction was performed according to protocol.
concentrations measured via NanoDrop:
- Sspl: 19,85 ng/µl
- PvuII: 10,57 ng/µl
Ligation:
For ligation, T4 ligase was used.
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
The Ligation was performed as following:
- SDM_SspI Volume: 4,03 µl
- SDM_PvuII Volume: 3,97 µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubating for 45 minutes.
Transformation:
Transformation performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol.
Design and ordering of primer for P40 Promoter BioBrick production
Investigator: Hanna
In order to generate a P40 promoter BioBrick = "Cap-Promoter", oligos were designed and ordered at Sigma Aldrich:
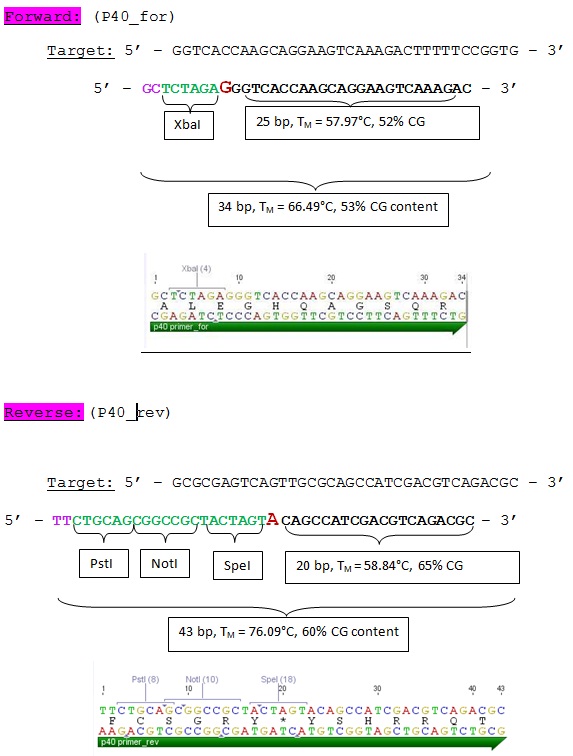
85. labday 10.08.2010
Retrafo with RepCap gene (Mr.Gene)
Investigator: Bea
Comments: We received the rep_cap insert ordered from Mr. Gene today. In order to used and clone the insert into the desired sequences a retrafo has to be performed.
Re-transformation:
- Construct used: pMA_REP_CAP
- We reveived 5 µg (lyophilzied). These were resuspended in 100 µL H2O and vortexted.
- c = 50 ng/µL
- c = Plasmid need to be diluted in order to obatain the final 500pg
- Dilution: 1:100
- 100 µL aliquot of XL1-B were thawed on ice. The proper amount of 1µL was added to the cells.
- Transformed cells were plated on LB plates containing ampicillin
Continuation: Cloning Rep40 & Rep52 into pSB1C3
A midi prep of the two Rep52+DMSO clones was performed yielding concentrations of 261,35 ng/µl (clone 1) and 240,47 ng/µl (clone 2). Unfortunately we noticed today that we should not use EcoRI to clone Rep52 into pSB1C3 because EcoRI cuts two times within the sequence of Rep52.
Midi-Prep of pAAV_RC, pAAV_RC_1.2 SDM SalI (P158), pAAV_RFC25_mGMK_TK30 clone 1 (P81) and 2 (P82)
Investigators: Patrick, Chris
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| pAAV_RC | P81a | P81b | P82a | P82b | P158a | P158b |
| 1348,49 ng/µl | 1783,86 ng/µl | 2096,25 ng/µl | 1915 ng/µl | 1469,35 ng/µl | 1834,25 ng/µl | 1771,27 ng/µl |
The annotations a and b were added because two midi-preps of P81, P82 and P158 were carried out.
Results of sequencing pSB1C3_pTAV2(P5) and Psb1C3_pAAV_RC(P5TATAless)
Investigator: Jessica
@Jessi: Which clones are ok, which not?? text does not correspond to pictures! (Bea)
sequencing of P180 was successful, P5 with TATAbox is in pSB1C3
sequencing of P183 was successful but there is a G missing --> plamid is thrown away
sequencing of P184 was successful, P5 is in without TATAbox. this plasmid can be used.
P181 and P180 are thrown away because of the result of the test digestion.
Repetition: BioBrick production of Rep40 and Rep 52
Investigator: Anna
Comments: PCR was repeated once again (that was bad!), because digestion was done with the wrong enzymes (EcoRI HF and SpeI). There are some differences in the PCR programm in comparison to the last (see 09.10.)
PCR
| Ingredients | Rep 40 | Rep52 |
| 5X Phusion HF buffer | 10 µl | 10 µl |
| 10 mM dNTP mix | 1µl | 1 µl |
| forward primer: O094 | 2,5µl | 2,5 µl |
| reverse primer: O0096/ O097 | 2,5 µl | 2,5 µl |
| DNA Template 1:100 (1ng) | 1 µl | 1 µl |
| DMSO | 1 µl | 1 µl |
| Phusion Polymerase | 0,5 µl | 0,5 µl |
| H2O | 31,5 µl | 31,5 µl |
| Total volume | 50 µl | 50 µl |
PCR programm
| PCR Program | temperature/ °C | Time | PCR products |
| 1 | 98 | 1min | |
| 2 | 98 | 15s | |
| 8x | *** | 15s | 62,5°C |
| 3 | 72 | *** | 25s |
| 4 | 98 | 15s | |
| 19x | *** | 15s | 67°C |
| 5 | 72 | *** | 25s |
| 6x | 72 | 5min | |
| Hold | 4 |
Agarose gel
0.5 g Agarose, 50 mL TAE (1 %),3 µL GELRED, at 120 Volt, running time: 45 minutes
| Sample | Sample / µl] | Loading dye (6x) / µl | Expected size |
|---|---|---|---|
| p22 (Rep40) | 50 µl | 8 µl | 940 bp |
| p23 (Rep52) | 50 µl | 8 µl | ~1200 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample Rep40 /µl | Sample Rep52 /µl | |
|---|---|---|---|
| Lane | 7 | 58 | 58 |
Comments: Digestion of the samples will be done tomorrow with XbaI and SpeI.
Gelextraction
Elution was done with 20 µl Pe buffer following the standard protocol.
c(Rep40)= 23,1 ng/µl
c(Rep52)= 78,0 ng/µl
Preparation of competent XL1blue and testtrafo
Investigator: Jessica
preparation of competent E.coli XL1blue was made according to the standardprotocol (91 eppis with 60µl) they are in the green boxes in -80°C
testtrafo was made
- one plate with 50pg/µl
- one plate with 100pg/µl
...trafo is failed because amp was missing on the plates (resistence in pUC). trafo will be repeated on aug 11th by Tobi...
Repetition: Mini-Prep and test digestion of pSB1C3_SDM_SspI/PvuII
Investigator: Adrian, Stefan
Plasmid Mini-Prep
- vector name: pSB1C3_SDM_SspI/PvuII
Glycerol Stocks
pSB1C3_SDM_SspI/PvuII
Only clone 1 grew, therefore there is just one stock = B159
Test digestion
- buffer used: 2
- Restriction-enzymes used:
- Enzyme 1 SspI
- Enzyme 2 PvuII
- Plasmid: pSB1C3_SDM_SspI/PvuII:
- Plasmid-Number: P185; DNA concentration: 47,9 ng/µL
- Plasmid-Number: P185; DNA concentration: 47,9 ng/µL
comment: DNA was eluted into previously used tube, therefore contamination is possible.
| Components | P185 Volume/µL | P51.1 Volume/µL |
| DNA | 18,1 | 6,8 |
| BSA (10x) | 2,5 | 2,5 |
| Buffer no. 2 (10x) | 2,5 | 2,5 |
| Enzyme 1 SspI | 0.5 | 0.5 |
| Enzyme 2 PvuII | 0.5 | 0.5 |
| H2O | 0,9 | 12,2 |
| Total volume | 10 | 10 | |
- Incubation: 45 min
Agarose-Gel:
0.4 g Agarose, 50 mL TAE, 3 µL GELRED, at 115 Volt, running time: 45 minutes
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P185 /µL | Sample P51.1 /µL | |
|---|---|---|---|
| Lane | 7 | 17 | 17 |
One cut is expected (PvuII in CFP).

Comment: There are 6 bands now. This test digestion again looks very different from that of P157.2 done by Jessica (lab day 82). Results should be discussed tomorrow.
Picking clones of pSB1C3_SDM_SspI/PvuII
Investigator: Stefan
Comments: Test digestion didn't work out, three additional clones were picked from the plate we get from yesterday's Trafo to retry tomorrow.
- Bacterial strain used: BL21
- Inoculating of 10 mL DYT medium containing 10µL chloramphenicol
- Put in 37°C room on shaker at 20.45hours.
86. labday 11.08.2010
Biobrick Assembly:Picking Clones, Miniprep, Testdigestion of CMV&lITR/ hGH & rITR
Investigator: Chris L., Achim
Test Digestion showed that the cloning was succesful.

NanoDrop concentrations:
- hGH_rITR1:105,15 ng/µl
- hGH_rITR2:96.97 ng/µl
- CMV_lITR1:231.61 ng/µl
- CMV_lITR2:129.19 ng/µl
Biobrick Assembly: bSB1C3_lITR_CMV & pSB1C3_betaglobin_mVenus
Investigator: Chris L., Achim
Comments: In order to proceed with the BioBrick assembly the plasmids pSB1C3_leftITR_CMV was cut with SpeI and PstI, pSB1C3_betaglobin_mVenus was cut with XbaI and PstI. The insert fragment containing betaglobin_mVenus was ligated to the vector fragment containing the leftITR_CMV promotor.
Digestion:
| components | pSB1C3_leftITR_CMV | pSB1C3_betaglobin_mVenus |
| DNA | 5,2 | 8,4 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme1 | 1 (SpeI) | 1 (Xba1) |
| Enzyme2 | 1 (PstI) | 1 (PstI) |
| H2O | 8,8 | 5,6 |
| Total volume | 20 | 20 |
Preparation of gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 45 minutes
Expected sizes of constructs:
- pSB1C3_leftITR_CMV: 2800 bp
- pSB1C3_betaglobin_mVenus: 1200 bp, 2000 bp
The corresponding bands were cut out and Gel-Extraction was performed according to protocol.
concentrations measured via NanoDrop:
- pSB1C3_leftITR_CMV: 55,5 ng/µl
- pSB1C3_betaglobin_mVenus:: 10,4 ng/µl
Continuation of Cloning of Rep40/52
Investigator: Anna
- Digestion of PCR products and vector:
| components | PCR product /µl | vector /µl |
| DNA | 19 | 10 |
| BSA (10x) | 3 | 3 |
| Buffer 4 (10x) | 3 | 3 |
| Enzyme XbaI | 1 | 1 |
| Enzyme SpeI | 1 | 1 |
| H2O | 3 | 12 |
| Total volume (e.g. 15,20,25,30 µl) | 30 | 30 |
Comments: This time digestion was done with XbaI and SpeI (means it has to be checked if the inserts are cloned into the vector in the right orientation).
- Purification of Rep40 and Rep52:
For the purification 95 µl of buffer PBI was used.
c(Rep40)= 12,5 ng/µl
c(Rep52)= 51,8 ng/µl
- Gelextraction of pSB1C3_RFC25_CFP:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 120 Volt, running time:55
5µl loading dye (6x) for the sample, Marker: GeneRuler ladder mix (Fermentas)
c(pSB1C3)= 10,59 ng/µl
c(pSB1C3_2)= 4,25 ng/µl
- Ligation of PCR products and vector:
For the Ligation 1µl T4 buffer (2x) and 1µl T4 ligase were used.
| Sample-no. | vector /µl | insert /µl | |
| pSB1C3 + Rep40 | 1 | 3,65 | 4,35 |
| pSB1C3 + Rep52 | 2 | 5,87 | 2,13 |
| Total volume (e.g. 15,20,25,30 µl) | 8 |
- Transformation:
The transformation was done following the standard protocol using B21 cells.
PCR of bla_14FM and ligation into pSB1C3_SDM_SspI and pSB1C3_SDM_PvuII for virobrick-production T
Investigator: Anissa,Kerstin
Comments: Because it still is unclear if SDM of pSB1C3_SspI_PvuII has worked, we decided to use the one time mutagenisised vector pSB1C3_SDM_SspI, to clone bla into it and to make the second mutation of PvuII after bla is inserted. .
- PCR:
(was performed following the standard protocol)
| Ingredients | v(pTB106_bla14FM) - DMSO / µl | v(pTB106_bla14FM) + DMSO / µl |
| 5X Phusion HF buffer | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 |
| forward primer: O116(P extreme f) | 2,5 | 2,5 |
| reverse primer: O117 (P extreme r) | 2,5 | 2,5 |
| DNA Template 1ng | 5,3 µl | 5,3 µl |
| DMSO (1%) | 0,5 | 0,5 |
| Phusion Polymerase | 0,5 | 0,5 |
| H2O | 28,2 | 27,7 |
| Total volume | 50 | 50 |
- PCR program:
| PCR Program | temperature/ °C |time | |
| 1 | 98 | 1' |
| 2 | 98 | 15 |
| 8x | 61 | 25 |
| 3 | 72 | 15 |
| 4 | 98 | 15 |
| 5 | 72 | 15 |
| 1x | 72 | 5' |
| Hold | 4 |
- Digestion PCR product:
- DNA-Concentration: 4,2 ng/µl
| components | volume / µl |
| DNA | 28,5 |
| BSA 100x | 0,4 |
| Buffer: 4 10x | 4 |
| Enzyme 1 (XbaI) | 1 |
| Enzyme 2 (PstI) | 1 |
| H2O | 5,1 |
| Total volume | 40 |
- Purification of PCR product: (QlAquick Gel Extraction Kit)
preparation of purification was made according to the standardprotocol
Comments: The wrong enzymes were used. Experiment has to be done tomorrow.
Test digestion pSB1C3_CFP_SDM SspI and PvuII
Investigator: Stefan
- buffer used: 2; Restriction-enzymes used: Enzyme 1: ClaI ; Enzyme 2: BpmI ; Enzyme 3: SspI ; Enzyme 4: PvuII
- Plasmids:
- pSB1C3_SDM_SspI/PvuII P185 (first tree lanes, cut with different enzymes)
- pSB1C3_RFC25_longlinker P105
- pSB1C3_CFP P51.1
| Components | P185/µL | P185/µL | P185/µL | P105/µL | P51.1/µL |
| DNA | 18 | 18 | 18 | 4,8 | 6,8 |
| BSA (10x) | 2,5 | 2,5 | 2,5 | 2,5 | 2,5 |
| Buffer 2 (10x) | - | - | - | 2,5 | 2,5 |
| Buffer 4 (10x) | 2,5 | 2,5 | 2,5 | - | - |
| Enzyme 1 ClaI (152) | 0,5 | 0,5 | 0,5 | - | - |
| Enzyme 2 BpmI (144) | 0,5 | 0,5 | 0,5 | - | - |
| Enzyme 3 SspI (68) | - | 0,5 | - | 0,5 | 0,5 |
| Enzyme 4 PvuII (50) | - | - | 1 | 0,5 | 0,5 |
| H2O | 1 | - | 0,5 | 14,2 | 12,2 |
| Total volume | 25 | 25 | 25 | 25 | 25 |
- Incubation: 75 minutes
Agarosegel
0.5 g Agarose, 50 ml TAE, 3 µL GELRED, at 115 Volt, running time: 60 minutes
comments: Test digestion of pSB1C3_SDM_SspI/PvuII with ClaI, BpmI and SspI looks the same as with ClaI and BpmI. Therefore we can assume that there is no additional restriction site within the vector. But still the bands above 3000 bp cannotz be explained. A new test digestion will be performed tomorrow to check the size of the vector.
The approach cut with ClaI, BpmI and PvuII looks strange. Propably, PvuII does not work correctly. New PvuII-HF was ordered and delivered so this can be verified tomorrow.
pSB1C3_RFC25_longlinker digested with SspI and PvuII seems to be not completely digested This could correspond to non-working PvuII.
pSB1C3_CFP looks like expected.
pSB1C3_SDM_Ssp1 (P125) sent for sequenzing
Investigator: Stefan
comment: To verify the correct deletion of the SspI restriction site it was sent for sequenzing.
Primer used: GATC_std_pTeSp-2
Picking clones of pSB1C3_SDM_SspI/PvuII (P185)
Investigator: Adrian
comment: pSB1C3_SDM_SspI/PvuII (P185) is empty. For plasmid production DYT + Cm was inoculated from the glycerol stock (B159).
87. labday 12.08.2010
XL1blue are ready to use
Investigator: Jessica
there are 60µl aliquots to use for one trafo
Mini-Prep of pSB1C3_SDM_SspI/PvuII and pMA_RC_insert
Investigator: Stefan
Gycerol stock was prepared for pMA_RC_insert and labeled B167. No glyerol stock for pSB1C3_SDM_SspI/PvuII because cells from earlier stock werer used for Mini-Prep.
Miniprep was performed according to protocol.
Samples were labeled:
- pSB1C3_SDM_SspI/PvuII = P185.1
- pMA_RC_insert = P190
Concentrations:
- pSB1C3_SDM_SspI/PvuII (P185.1): 257,2 ng/µl
- pMA_RC_insert (P190): 339,5 ng/µl
No test digestion was performed.
Biobrick Assembly: pSB1C3_lITR_CMV & pSB1C3_betaglobin_mVenus
Investigator: Chris L., Achim
Comments: In order to proceed with the BioBrick assembly the plasmids pSB1C3_leftITR_CMV was cut with SpeI and PstI, pSB1C3_betaglobin_mVenus was cut with XbaI and PstI. The insert fragment containing betaglobin_mVenus was ligated to the vector fragment containing the leftITR_CMV promotor.
Ligation:
For ligation, T4 ligase was used.
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Ligation was carried out as following:
- pSB1C3_leftITR_CMV: 6,98 µl
- pSB1C3_betaglobin_mVenus: 1,02 µl
Incubation for 40 minutes.
- Transformation:
The transformation was done following the standard protocol using XL1B cells.
Repetition of cloning of bla14FM_viralbrick_MCS into pSB1C3_SDM_SspI
Investigator: Anissa
Comments:Yesterday the pcr-product of bla14FM was cut with the wrong enzymes, so no overhangs could result. Today it will be repeated with the rihgt ones (SpeI and NgomIV) .
- Digestion of the PCR product bla14FM for 2 h :
- DNA-Concentration: 10,2 ng/µl
| components | volume / µl |
| DNA | 18 |
| BSA 100x | 3 |
| Buffer: 4 10x | 3 |
| Enzyme 1 (NgomIV) | 1 |
| Enzyme 2 (SpeI) | 1 |
| H2O | 4 |
| Total volume | 30 |
- Purification of PCR product: (QlAquick Gel Extraction Kit)
preparation of purification was made according to the standardprotocol
- Digestion of vector backbone pSB1C3_SDM_SspI for 2 h:
- 1 µg vector has been used
| components | volume / µl |
| DNA | 3,12 |
| BSA 100x | 1,5 |
| Buffer: 4 10x | 1,5 |
| Enzyme 1 (NgomIV) | 1 |
| Enzyme 2 (SpeI) | 1 |
| H2O | 6,88 |
| Total volume | 15 |
- the cut vector was loaded on a preperative 1% agarose gel and the gel was running for 45 minutes
- gel was cut and gelextraction was performed according the qiagen standard protocol.
- Ligation of pSB1C3_SDM_SspI and bla14FM:
| components | volume / µl |
| t4 buffer (10X) | 1 |
| t4 Ligase | 1 |
| vector DNA | 2,72 |
| Insert | 5,28 |
- Transformation: pSB1C3_SDM_SspI_bla14FM was transformed into BL21 following the stanard protocol
Quickchange of the EaglII restriction site in the PMA_RepCap vector
Investigator: Chris L., Achim
Comments: We performed a QuikChange mutagenesis on the synthesized PMA_RepCap vector (P190). The EaglII restriction enzyme also cuts NotI (IGEM standard) therefore EaglII has to be changed into a different recognition site. Primers were designed to change it into PvuII. The new QuikChange Lighting Site-Directed Mutagenesis Kit from Stratagene was used, therefore the mutagenesis was carried out as described in the QuikChange Manual. The PCR product was transformed into XL10-Gold cells, we used the IGEM standard protocol, except for the addition of ß-ME to the XL10-Gold cells. :
PCR-reaction:
| Volume / µl | ingredients | recommended /µl |
| 2,5 | 10X reaction buffer | 2,5 |
| 2,9 | P190 template (~10 ng) | 2,9 of 1:100 dilution |
| 0,51 | forward primer: O122 | 62,5 ng |
| 0,51 | reverse primer: O123 | 62,5 ng |
| 0,5 | dNTP | 250 µM each dNTP |
| 17,6 | H2O | |
| 0,5 | QuikChange Lightning ennzyme(1.25) |
PCR program:
| Rounds | temperature/ °C | Time |
| 1 | 95 | 2 minutes |
| 18 | 95 | 20 seconds |
| 18 | 60 | 10 seconds |
| 18 | 68 | 2 minutes (4 kb * 30 sec) |
| 1 | 68 | 5 minutes |
The PCR product was digestet with Dpn I for 10 minutes at 37°C
Plasmid was transformed into XL10-Gold cells.
Sequencing results of Rep68, Rep78 and AAP BioBricks
Investigator: Hanna
Comment: pSB1C3_Rep78 (P160), pSB1C3_Rep68 (P165) and pSB1C3_AAP (P170) were sent for sequencing twice. No sequencing made sense. Paradoxically the Rep78 sequencing result fit to the p5 promoter BioBrick...
By having a closer look to the BioBrick PCR we found out that the fragment sizes didn't fit to the expected sequences. In addition to that the test digestion also showed some discrepancies.
Because we weren't able to find out in which steps of the BioBrick production things went wrong, we decided to discard all referring glycerol stocks and plasmids and to perform the Rep68, Rep78 and AAP BioBrick PCR again!!!
Comment 13.08.2010 (Hanna): We wondered how the whole Rep stuff could be sequenced reverse AND forward without any forward sequencing primer... By checking the "sequencing book" we found out that always the wrong primers were used: the GOI primers can be used in order to sequence the Gene of interest = GOI (!) in pAAV_MCS and NOT the pSB1C3!!!!!!!!!
Order oligos for CD-biobrick
Investigator: Jessica and Kira
ordered on aug 12th
BioBrick assembly of pSB1C3_leftITR_CMV_mVenus
Investigator: Stefan
comment: In order to assemble our complete constructs mVenus has to be included into the pSB1C3_leftITR_CMV vector.
Digestion:
| components: | pSB1C3_leftITR_CMV P188(Vector) | pGA14_mVenus P60 (Insert) |
| DNA | 6 | 8 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme1 | 1 (SpeI) | 1 (Xba1) |
| Enzyme2 | 1 (PstI) | 1 (PstI) |
| H2O | 8 | 6 |
| Total volume | 20 | 20 |
Preparation of gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 110 Volt, running time: 45 minutes
Expected sizes of constructs:
- pSB1C3_leftITR_CMV: 2869 bp
- pGA14_mVenus: 2896 bp, 756 bp
The corresponding bands were cut out and Gel-Extraction was performed according to protocol.
concentrations measured via NanoDrop:
- pSB1C3_leftITR_CMV: 39,2 ng/µl
- pGA14_mVenus: 8,2 ng/µl
Ligation:
T4 ligation was performed according to protocol.
Volumes used:
- vector: 1,67 ng/µl
- insert: 6,33 ng/µl
Transformation:
bacterial strain used: XL1bB
antibiotic used for plate: chlorampenicol
Transformation was performed according to protocol.
Plate was prepared and put in 37°C room.
Opened a BioBrick box
Investigator: Bea
In order to "save" the plasmids/BioBricks which has been prepared and confirmed for submitting it to the parts registry a new box have been prepared. The box should contain 30 µL of the desired construct cloned into the pSB1C3 vector and the BBa number given from the headquarter.
The first BioBricks which are in this box are:
- pSB1C3_hGH (P136): Bba_K404002
- pSB1C3_betaglobin (P133: Bba_K404001
- pSB1C3_CMV (P145): Bba_K404003
- pSB1C3_leftITR (P175): Bba_K404006
- pSB1C3_rightITR (P172): Bba_K404005
- pSB1C3_mGMK (P156): Bba_K404004
In our nomenclature plasmid excel list the constructs which we aliquoted should be highlighted in blue and commented with the given Bba number. Additionally it should be taken in consideration that enough DNA should be in the "working" plasmid box. Therefore, if necessary, inoculate DYT medium for Mini-Preps.
In this case ALL the glycerol stocks were used for incoluating 10 mL DYT medium containing 10 µL chloramphenicol respectively.
To do: Mini-Preps of the over night cultures.
88. labday 13.08.2010
Continuation:pSB1C3_SDM_SspI_bla14FM
Investigator: Patrick
Intention: receive enough plasmid for a sequencing and maybe futher working steps.
Three clones were picked in the morning to inoculate DYT and perform a mini-prep in the evening and to send them for sequencing. The three clones were picked and prepped according to the standard protocol yielding the following DNA concentrations:
- pSB1C3_SDM_SspI_bla14FM clone 1: 65,15 ng/µl , Sequencing labeled: PS3.1
- pSB1C3_SDM_SspI_bla14FM clone 2: 22,3 ng/µl , Sequencing labeled: PS3.2
- pSB1C3_SDM_SspI_bla14FM clone 3: 62,3 ng/µl , Sequencing labeled: PS3.3
Used primer: seq_pSB1C3_VR2_rev (O51)
Results: see http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal#Sequence_analysis_of_pSB1C3_SDM_SspI_BLA
Mini-preps of pSB1C3_hgh,pSB1C3_beta globin,pSB1C3_left ITR,pSB1C3_right ITR,pSB1C3_mGMK,pSB1C3_CMV
Comments:new mini-preps of the glycerol-stocks ofpSB1C3_hgh(=B103),pSB1C3_beta globin(=B100),pSB1C3_left ITR (=B143),pSB1C3_right ITR (=B140),pSB1C3_mGMK (=B125),pSB1C3_CMV :
| components | concentration in ng/µl |
| P191 (pSB1C3_right ITR) | 100,74 |
| P192 (pSB1C3_beta-globin) | 240,08 |
| P193 (pSB1C3_CMV) | 333,89 |
| P194 (pSB1C3_hgh) | 328,42 |
| P195 (pSB1C3_mGMK) | 258,63 |
| P196 (pSB1C3_left ITR) | 173,98 |
Design and ordering of oligos for VP fusion targeting approaches
Investigator: Hanna
Comment: In order to target the EGFR receptor of our A431 cells different approaches have been figured out: Besides loop insertions, N-terminal fusion to the VP2 protein will be performed. In addition to that a VP1 insertion approach (after Grieger et al., 2007) was planned.
Fusion and insertion molecules will be e.g. His-tag, Affibody, Darpin, GFP. These approaches can be combinated with the loop insertions.
For this purpose the following oligos were ordered today:
Mini-Prep and Test digestion of Rep40 and Rep52 of the cloning experiment ("Cloning of Rep40/52" 11.08.2010 Investigator: Anna)
Investigator: Chris L., Achim
Comments: There are small and big colonies on the plate. Maybe problems with the agar plate. Picked colonies of pSB1C3_Rep40 sample 1 and pSB1C3_Rep52 sample 1 are small colonies. Picked colonies of pSB1C3_Rep40 sample 2 + 3 and pSB1C3_Rep52 sample 2 + 3 are standard size colonies.
Nanodrop
- Rep 40 Sample 1: 156,26 ng/µl P197
- Rep 40 Sample 2: 200,05 ng/µl P198
- Rep 40 Sample 3: 144,06 ng/µl P199
- Rep 52 Sample 1: 173,36 ng/µl P200
- Rep 52 Sample 2: 160,75 ng/µl P201
- Rep 52 Sample 3: 295,77 ng/µl P202
Test digestion 800ng DNA
| components | P145 /µl | P146 /µl | P145 /µl | P146 /µl | P145 /µl | P146 /µl |
| DNA | 5,1 | 4 | 5,6 | 4,6 | 5 | 2,7 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1: XbaI | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| Enzyme 2: SpeI | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 9,9 | 11 | 9,4 | 10,4 | 10 | 12,3 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 | 20 | 20 | 20 |
Incubation time: 45 minutes, Incubation temperature: 37°
0,5 g Agarose, 50ml x TAE, 3µl GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| Rep 40 1 | 20 µl | 4 µl | ____ bp | 940 bp |
| Rep 40 2 | 20 µl | 4 µl | ____ bp | 940 bp |
| Rep 40 3 | 20 µl | 4 µl | ____ bp | 940 bp |
| Rep 52 1 | 20 µl | 4 µl | ____ bp | 1198 bp |
| Rep 52 2 | 20 µl | 4 µl | ____ bp | 1198 bp |
| Rep 52 3 | 20 µl | 4 µl | ____ bp | 1198 bp |
| Marker | Sample Rep40 1 /20µl | Sample Rep40 2 /20µl | Sample Rep40 3 /20µl | Sample Rep52 1 /20µl | Sample Rep52 2 /20µl | Sample Rep52 3 /20µl | |
|---|---|---|---|---|---|---|---|
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
agarose gel: 1%, digestion: 45 minutes, 37°C
Comments:Results look bad. Rep52 clone 3 looks good, send for sequencing.
</p>
Repetition of Rep 40 and Rep 52 ligation and transformation with the vector pSB1C3_CFP
Investigator: Chris L.
- Ligation of PCR products and vector:
For the Ligation 1µl T4 buffer (10x) and 1µl T4 ligase were used.
| Sample-no. | vector /µl | insert /µl | |
| pSB1C3 + Rep40 | 1 | 4,92 | 3,08 |
| pSB1C3 + Rep52 | 2 | 6,47 | 1,53 |
| Total volume (e.g. 15,20,25,30 µl) | 10 | 10 |
- Transformation:
The transformation was done following the standard protocol using XL1B cells.
Strategy for VP2 N-terminal fusion and VP1 insertion of targeting moelcules
Investigator: Hanna
Comment: In order to target the EGFR receptor of tumor cells via Affibody or Darpin (antibody fragment is also possible!!!), or in order to expose e.g. His-Tags on the virus surface, two approaches have been figured out.


pSB1C3_Rep52 (P202) sent for sequencing
Investigator: Chris L.
Primer used: Reverse Primer VR2
<p style="font-size:15px; background-color:#66bbff;">Repetition: Biobrick production of Rep 68, 78 and AAP
Investigator: Stefan
Aim of the experiment:
We want to produce biobricks from Rep 68, Rep 78 and the AAP.
- Plasmids used as template:
Rep_68_ex (p119): c = 470,6 ng/µl
Rep_78_(p122): c = 566,8 ng/µl
pAAV-RC containing AAP ORF (p50): c = 378,5 ng/µl
- Primer used:
For Rep_68_ex: Praefix_68_78_ex & Suffix_40_68_ex
For Rep_78_ex: Praefix_68_78_ex & Suffix_52_78_ex
For AAP_ex: Praefix_AAP_ex & Suffix_AAP_ex
- PCR:
(was performed following the standard protocol)
| Ingredients | Volume / µl | Rep68 | Rep78 | AAP |
| 5X Phusion HF buffer | 10 | |||
| 10 mM dNTP mix | 1 | |||
| forward primer: | 2,5 | |||
| reverse primer: | 2,5 | |||
| DNA Template | *** | 2,5 µl | 2 µl | 3 µl |
| DMSO (2%) | - | - | 1 µl | |
| Phusion Polymerase | 0,5 | |||
| H2O | *** | 31 µl | 31,5 µl | 29,5 µl |
| Total volume | 50 |
PCR program:
| PCR Program | temperature/ °C | Time | Rep68 | Rep78 | AAP |
| 1 | 98 | 1min | |||
| 2 | 98 | 15s | |||
| 3 | *** | 25s | 63°C | 62°C | 64°C |
| 4 (step 2-4 8x) | 72 | *** | 24s | 27s | 10s |
| 5 | 98 | 15s | |||
| 6 | *** | 25s | 68°C | 64°C | 68°C |
| 7 | 72 | *** | 24s | 27s | 10s |
| 8 (step 6-8 17x) | 72 | 5min | |||
| Hold | 4 |
Used agarose gel: 0,5 g Agarose,50 ml TBE (1%), 3 µl GELRED , at 115 Volt, running time:45 minutes
Gel extraction
Gel extraction was perfomed according to protocol.
- Digestion of PCR products and vector:
| components | PCR products /µl | vector /µl |
| DNA | 29 | 11 |
| BSA (10x) | 4 | 2 |
| Buffer 4 (10x) | 4 | 2 |
| Enzyme XbaI | 1 | 1 |
| Enzyme SpeI | 1 | 1 |
| H2O | 1 | 3 |
| Total volume (e.g. 15,20,25,30 µl) | 40 | 20 |
Comments: Digestion was done with XbaI and SpeI, it has to be checked if the inserts are cloned into the vector in the right orientation.
- Purification of Rep68, 78 and AAP:
For the purification 200 µl of buffer PBI was used.
c(Rep68)= 14,3 ng/µl
c(Rep78)= 12,0 ng/µl
c(AAP)= 25,1 ng/µl
- Gelextraction of pSB1C3_RFC25_CFP:
0,5 g Agarose,50 ml TBE (1%), 3 µl GELRED , at 115 Volt, running time:50 minutes
4µl loading dye (6x) for the sample, Marker: GeneRuler ladder mix (Fermentas)
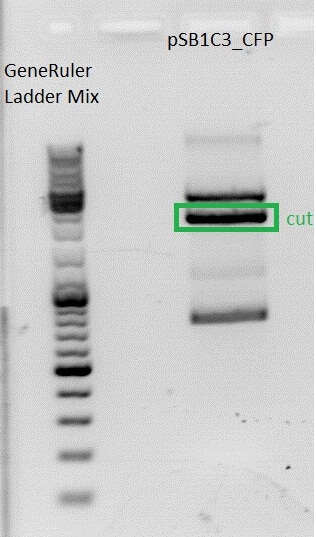
c(pSB1C3)= 6,9 ng/µl
- T4 ligation of PCR products and vector:
For the Ligation 1µl buffer (10x) and 1µl T4 ligase were used.
| vector /µl | insert /µl | |
| pSB1C3 + Rep68 | 3,78 | 4,24 |
| pSB1C32 + Rep78 | 3,13 | 4,87 |
| pSB1C3 + AAP | 6,42 | 1,58 |
- Transformation:
The transformation was done following the standard protocol using XL1b cells.
89. labday 14.08.2010
Sequence analysis of pSB1C3_SDM_SspI_BLA
Investigator: Patrick, Bea
Comments: Sequence analysis of three clones revealed that all sequences contain some mutations in the regions where the primer annealed. This was due to the extreme primer lentgh. Primers that long should be ordered as HPLC purified in order to avoid mutations in the primer sequence.
- Clone 1: Mutation in the SalI site of the loop insertion site in the Viral Brick MCS lead to the removal of the recognition site. CANNOT be used for the loop insertions!!!!
- Clone 2: Mutation in the region of the primer upstream of the loop insertion sites SalI and PvuII.
- Clone 3: Mutation in the region of the primer downstream of the loop insertion sites SalI and PvuII.
Next steps: Two additional clones will be picked and sent for sequencing (clones 4 and 5). In order to obtain a construct without any mutations, clone 2 and clone 3 couldbe cloned together. Each clone will be digested with SalI and BamHI. Clone 2 will be the "insert", clone 3 is going to be the "vector". Therefore the fragments will (hopefully) not contain anymore mutations in the relevant sequence.
Mini Preps of PMA_RepCap vector and pSB1C3_left ITR_CMV_betaglobin_mVenus
Investigator: Chris L., Bea
Comments:new mini-preps of the glycerol-stocks of pSB1C3_leftITR_CMV_betaglobin_mVenus clone 1 (=B179), pSB1C3_leftITR_CMV_betaglobin_mVenus clone 2 (=B180), pSB1C3_leftITR_CMV_betaglobin_mVenus clone 3 (=B181), pMA_RepCap Vector_SDM_InsPvuII clone 1, (=B182), pMA_RepCap Vector_SDM_InsPvuII clone 2 (=B183) and pMA_RepCap Vector_SDM_InsPvuII clone 3 (=B184)
Comment: The plamsid numbers in the picture (P179-P184) correspond to the glycerol stocks B179-B184. The corresponding plasmid numbers are: P208-P213.
Nanodrop
- pSB1C3_leftITR_CMV_betaglobin_mVenus clone 1: 256,64 ng/µl B179
- pSB1C3_leftITR_CMV_betaglobin_mVenus clone 2: 274,86 ng/µl B180
- pSB1C3_leftITR_CMV_betaglobin_mVenus clone 3: 217,20 ng/µl B181
- pMA_RepCap Vector_SDM_InsPvuII clone 1: 253,71 ng/µl B182
- pMA_RepCap Vector_SDM_InsPvuII clone 2: 195,64 ng/µl B183
- pMA_RepCap Vector_SDM_InsPvuII clone 3: 144,61 ng/µl B184
Test digestion 800ng DNA
| components | B179 /µl | B180 /µl | B181 /µl | B182 /µl | B183 /µl | B184 /µl | pMA_RepCap Vector /µl |
| DNA | 3,1 | 2,9 | 3,7 | 3,1 | 4 | 5,5 | 2,4 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1: NgoMIV | 0,5 | 0,5 | 0,5 | 0 | 0 | 0 | 0 |
| Enzyme 2: EcoRI | 0,5 | 0,5 | 0,5 | 0 | 0 | 0 | 0 |
| Enzyme 2: PvuII | 0 | 0 | 0 | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 11,9 | 12,1 | 11,3 | 12,4 | 11,5 | 10 | 13,1 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
Incubation time: 45 minutes, Incubation temperature: 37°
0,5 g Agarose, 50ml x TAE, 3µl GELRED, at 115 Volt, running time: 45 minutes
Results of test digestion
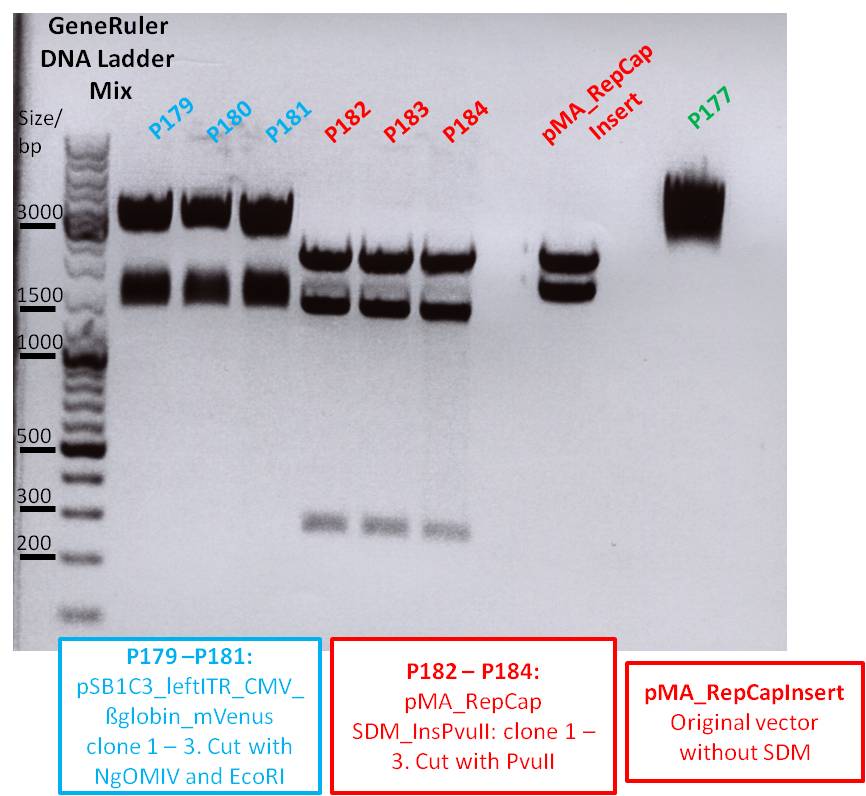
Results: Two (three) different approaches can be seen on the agarose gel.
- P178-P181 (blue): The plasmid pSB1C3_leftITR_CMV_betaglobin_mVenus was digested with NgoMIV and EcoRI. Expected bands were: 2873bp and 1343 . As it can be seen in the picture, the two fragments ran too high (~300bp). As it is almost normal for the pSB1C3 plasmid, we should figure out why this is always the problem for this vector.
- P182-P183 (red): The plasmid pMA_RepCap_SDM_insPvuII was digested with PvuII as well as the control vetor pMA_RepCap where no site-directed mutagenesis was performed in order to see the difference. As it can be seen in the picture above, the expected band sizes of 263bp,1384bp and 2133bp correspond to the bands detected in the gel. The control vector instead reveals only two bands detectable. Therefore site-directed mutagenesis was successfully performed.
- P177 (green): pSB1C3_hTERT. For further details see extra part below (Test digestion of pSB1C3_hTERT.
Mini-Prep of pSB1C3_leftITR_CMV_mVenus
Investigator: Stefan
Glycerol stocks were prepared:
Test digestion: or test digestion, pSB1C3_leftITR_CMV (P188) was digested as well.
| Ingredients | clone 1/µl | clone 2/µl | P188/µl |
| DNA: | 3,5 | 3,5 | 4 |
| BSA (10x): | 2 | 2 | 2 |
| Buffer 4: | 2 | 2 | 2 |
| Enzyme 1 EcoRI: | 0,5 | 0,5 | 0,5 |
| Enzyme 2 AgeI | 0,5 | 0,5 | 0,5 |
| H2O | 6,5 | 6,5 | 6 |
-
All samples were loaded on a 1% agarose gel:
- GeneRuler Ladder Mix (7µl)
- P206 = pSB1C3_leftITR_CMV_mVenus clone 1
- P207 = pSB1C3_leftITR_CMV_mVenus clone 2
- P188 = pSB1C3_leftITR_CMV
comment: Through digestion, fragments would have similar sizes. Therefore, a new digestion will be performed tomorrow.
Test digestion of pSB1C3_pHTERT
Investigator: Bea
Comments: pSB1C3_hTERT was sent for sequencing and revealed that the PCR and the inserting of the PCR product : promoter for hTERT was sucessfully performed. But the sequencing results showed a new AgeI restriciton site in the plasmid backbone. We could not see if the new recognition site was due to the bad sequence read or that there is really a new AgeI site. Therefore, a test digestion will be perfomed with PstI and AgeI in order to figure our wheter AgeI is present in the backbone or not.
Sequence analysis of pSB1C3_hTERT:
First the sequenced plasmid P177 was digested:
| P177/µL | |
| DNA | 3 |
| BSA (10x) | 1,5 |
| Buffer 4 (10x) | 1,5 |
| Enzyme PstI | 0,5 |
| Enzyme AgeI | 0,5 |
| H2O | 8 |
| Total volume | 15 |
- Incubation time: 45minutes
- Incubation tempereature: 37°C
After the incubation,the samples were loaded on the 1% agarose gel, and run at 110 V for 45 minutes.
Result of the test digestion:
Results: As it can be seen in the sequencing results of pSB1C3_hTERT there could have been an additional AgeI site in the vector. If AgeI would have been in the vector, two fragments have been expected by digetsing the plasmid with AgeI and PstI. As it can be seen in the agarose gel, lane 11 (labeled with the green P177) only reveals one lane instead of two. Therefore it can be assumed that NO AgeI site is in the pSB1C3 backbone.
Picking clones ...
Investigator: Patrick 15 Clones were picked saturday night so that a mini-prep could be performed on sunday.
90. labday 15.08.2010
Sequence analysis of pSB1C3_Rep52
Investigator: Bea
Comments: Clone 3 was sent for sequencing because this clone revealed the right band sizes of the test digestion performed friday. For more details see: [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal#Mini-Prep_and_Test_digestion_of_Rep40_and_Rep52_of_the_cloning_experiment_.28.22Cloning_of_Rep40.2F52.22_11.08.2010_Investigator:_Anna.29 Test digestion]
Sequence assembly showed that the suffix of the Rep protein 52 is ok and that PCR worked well.
There can be seen that a silent mutation from AGG to AGA was inserted, but it will have no effect on the protein because codon usage in H. sapiens is nearly the same for both codons.
The prefix could not be analysed because of bad sequence read quality. For further verification, another sequencing with the forward primer has to be performed.
Next steps: Send plasmid for another sequencing round with forward primer in order to analyse the prefix.
[http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]
Mini-Prep of pSB1C3_Rep68, pSB1C3_Rep78, pSB1C3_RepAAP, pSB1C3_Rep40, pSB1C3_Rep52
Investigator: Stefan
-
Glycerol stocks were prepared:
- B185 = pSB1C3_Rep68 clone 1
- B186 = pSB1C3_Rep68 clone 2
- B187 = pSB1C3_Rep68 clone 3
- B188 = pSB1C3_Rep78 clone 1
- B189 = pSB1C3_Rep78 clone 2
- B190 = pSB1C3_Rep78 clone 3
- B191 = pSB1C3_AAP clone 1
- B192 = pSB1C3_AAP clone 2
- B193 = pSB1C3_AAP clone 3
- B194 = pSB1C3_Rep40 clone 1
- B195 = pSB1C3_Rep40 clone 2
- B196 = pSB1C3_Rep52 clone 1
- B197 = pSB1C3_Rep52 clone 2
- B198 = pSB1C3_SDM_SspI_Bla14FM clone 4
- B199 = pSB1C3_SDM_SspI_Bla14FM clone 5
Mini-Prep following the standard protokol
- P208 = pSB1C3_Rep68 clone 1 c=230,0
- P209 = pSB1C3_Rep68 clone 2 c=160,5
- P210 = pSB1C3_Rep68 clone 3 c=128,6
- P211 = pSB1C3_Rep78 clone 1 c=150,8
- P212 = pSB1C3_Rep78 clone 2 c=108,6
- P213 = pSB1C3_Rep78 clone 3 c=131,3
- P214 = pSB1C3_AAP clone 1 c=142,1
- P215 = pSB1C3_AAP clone 2 c=148,3
- P216 = pSB1C3_AAP clone 3 c=163,0
- P217 = pSB1C3_Rep40 clone 1 c=164,5
- P218 = pSB1C3_Rep40 clone 2 c=<145,1br />
- P219 = pSB1C3_Rep52 clone 1 c=156,0
- P220 = pSB1C3_Rep52 clone 2 c=164,0
- P221 = pSB1C3_SDM_SspI_Bla14FM clone 4 c=220,4
- P222 = pSB1C3_SDM_SspI_Bla14FM clone 5 c=252,2
Test digestion:
No test digestion for pSB1C3_SDM_SspI_Bla14FM clone 4 and 5.
For test digestion, pSB1C3_leftITR_CMV_mVenus (P206-207) and pSB1C3_leftITR_CMV (P188) was digested as well.
Samples P208-220 were digested with XbaI and SpeI.
Samples P207, P207 and P188 were digested with XbaI and NgoMIV.
Gel used:
100 ml TAE (1x), 1 g agarose, 6 µl GELRED
115 V, 50 minutes
| Ingredients | P208/µl | P209/µl | P210/µl | P211/µl | P212/µl | P213/µl | P214/µl | P215/µl | P216/µl | P217/µl | P218/µl | P219/µl | P220/µl | P206/µl | P207/µl | P188/µl |
| DNA: | 4 | 6 | 7 | 6 | 9 | 7 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 3,5 | 3,5 | 4 |
| BSA (10x): | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Buffer 4: | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| Enzyme 2 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 11 | 9 | 8 | 9 | 6 | 8 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 11,5 | 11,5 | 11 |

comment: Rep- and AAP approaches have to be repeated. We did not realize that the vector can religate! Plasmids and glycerol stocks were thrown away.
Inoculation of CD colony
Investigator: Kira
One colony was picked from agar plate and dispersed into 10 ml DYT+ Amp and incubated over night @ 37 C.
 "
"