|
AUGUST: WEEK 2
August, 9th
Phasins plates showed very few colonies (12 for I20-new and 17 for I21-new): they were all picked and let grow in LB+Amp 100ug/ml. In the evening we made glycerol stocks and re-filled the tubes for the screening of the following day:
| I20-new: | I20-1 | I20-2 | I20-3 | I20-4 | I20-5 | I20-6 | I20-7 | I20-8 | I20-9 | I20-10 | I20-11 | I20-12
|
| I21-new: | I21-1 | I21-2 | I21-3 | I21-4 | I21-5 | I21-6 | I21-7 | I21-8 | I21-9 | I21-10 | I21-11 | I21-12 | I21-13 | I21-14 | I21-15 | I21-16 | I21-17
|
In order not to loose our time we decided to perform again the PCR-amplification/modification of <partinfo>BBa_K208001</partinfo> in case of a negative screening. Than we gel-ran and gel-extracted right amplicons.
PCR was performed with:
- 10_F and S_R primers
- S_F and S_R primers
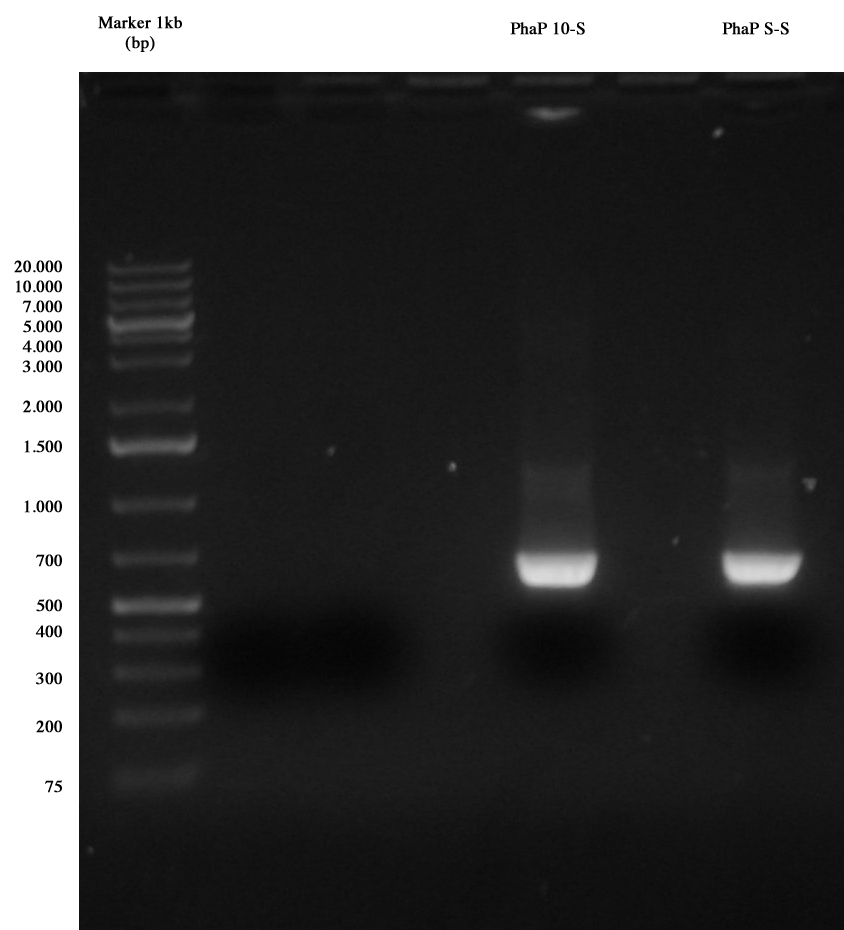 PCR results: PhaP 10-S and PhaP S-S The image shows results of our PCR: both amplicons are positive, so we purified cut DNA, obtaining the following quantifications:
- PhaP 10-S: 16,8 ng/ul
- PhaP S-S: 17,1 ng/ul
Tomorrow we will proceed to digest this part, in order to have an alternative if all clones of I20 and I21 are negative.
In the afternoon, we inoculated all parts we will need tomorrow.
| Part | Medium | Task
|
| <partinfo>BBa_B0034</partinfo> | 5ml LB+Amp | Ligation
|
| I15-1 | 5ml LB+Amp | Ligation
|
| Linker (<partinfo>BBa_K105012</partinfo>) | 5ml LB+Amp | Ligation
|
| <partinfo>pSB4C5</partinfo> | 5ml LB+Cm 12,5 | Ligation
|
| I22-1 | 5ml LB+ Amp (picked from colony) | Ligation/Screening/Glycerol stock
|
| I22-2 | 5ml LB+ Amp (picked from colony) | Ligation/Screening/Glycerol stock
|
| I22-3 | 5ml LB+ Amp (picked from colony) | Ligation/Screening/Glycerol stock
|
| I23-1 | 5ml LB+ Amp (picked from colony) | Screening/Glycerol stock
|
| I23-2 | 5ml LB+ Amp (picked from colony) | Screening/Glycerol stock
|
| I23-3 | 5ml LB+ Amp (picked from colony) | Screening/Glycerol stock
|
| <partinfo>BBa_r0062</partinfo> | 5ml LB+ Amp | Ligation
|
| I7_4C5-2 | 1ml LB+ Cm12,5 | Tecan Test
|
| I8_4C5-2 | 1ml LB+ Cm12,5 | Tecan Test
|
| I10_4C5-1 | 1ml LB+ Cm12,5 | Tecan Test
|
| I12_4C5-1 | 1ml LB+ Cm12,5 | Tecan Test
|
| A2(=<partinfo>BBa_J23100</partinfo>+GFP) | 1ml LB+ Amp | Tecan Test
|
| <partinfo>BBa_B0032</partinfo> | 1ml LB+ Amp | Tecan Test
|
| I8-5 D | 1ml LB+ Amp (picked from colony) | Tecan Test/Glycerol stock
|
| I8-5 E | 1ml LB+ Amp (picked from colony) | Tecan Test/Glycerol stock
|
| I8-5 F | 1ml LB+ Amp (picked from colony) | Tecan Test/Glycerol stock
|
Check of LB+Cm 6 ug/ml agar plates. We let grow at 30°C:
- MG1655 in LB
- MC1061 in LB
- MG123/008 in LB+Amp 50 ug/ml
- MC123/008 in LB+Amp 50 ug/ml
- F2620-4C5 (positive control)
Than we streaked cultures on a seven-sector divided plate, and we let it grow ON at 30°C. Since 6 ug/ml is a very low concentration, we wanted to check if actually nothing (except F2620-4C5) grow on these plates.
Transformation at 30°C of 100 ul of MG123/008 and MC123/008 competent cells to check their efficiency. We used 1 ul (~4 ng) of RING, F2620-4C5, NOTHING (water) and plated on proper LB agar plates:
- RING: Cm 34 ug/ml
- F2620-4C5 (positive control): Cm 12,5 ug/ml
- NOTHING (negative control): Cm 12,5 ug/ml
Both F2620-4C5 and RING should survive (not RING when in XX123), but we had a problem so that MC008 transformed with RING couldn't be plated. We will check it another time. We let grow plates ON, 30°C.
August, 10th
Glycerol stock was prepared for:
- linker (<partinfo>BBa_K105012</partinfo>)
- I22-1
- I22-2
- I22-3
- I23-1
- I23-2
- I23-3
- I8-5 D
- I8-5 E
- I8-5 F
and are stored at -80°C.
Cultures were diluted (5ul in 2ml LB+antibiotic) for TECAN test:
| I8-5 D | I8-5 E | I8-5 F
|
| I74C5 | I84C5 | I104C5
|
| I124C5 | Rbs32 | A2
|
MiniPrep was performed for following cultures (using our new NucleoSpin kit, that we prepared yesterday! :) ):
| Culture | Quantification
|
| I20-1 | 16,5 ng/ul
|
| I20-2 | 8,9 ng/ul
|
| I20-3 | 19,3 ng/ul
|
| I20-4 | 12,5 ng/ul
|
| I20-5 | 22,4 ng/ul
|
| I20-6 | 12,3 ng/ul
|
| I20-7 | 4,1 ng/ul
|
| I20-8 | 15,7 ng/ul
|
| I20-9 | 14,4 ng/ul
|
| I20-10 | 17,5 ng/ul
|
| I20-11 | 24,5 ng/ul
|
| I20-12 | 21,2 ng/ul
|
| I21-1 | 22,2 ng/ul
|
| I21-2 | 24,6 ng/ul
|
| I21-3 | 14,2 ng/ul
|
| I21-4 | 12,4 ng/ul
|
| I21-5 | 19,4 ng/ul
|
| I21-6 | 17,0 ng/ul
|
| I21-7 | 14,5 ng/ul
|
| I21-8 | 23,1 ng/ul
|
| I21-9 | 18,9 ng/ul
|
| I21-10 | 16,4 ng/ul
|
| I21-11 | 13,3 ng/ul
|
| I21-12 | 15,5 ng/ul
|
| I21-13 | 33,5 ng/ul
|
| I21-14 | 22,2 ng/ul
|
| I21-15 | 21,1 ng/ul
|
| I21-16 | 23,8 ng/ul
|
| I21-17 | 20,6 ng/ul
|
| I22-1 | 117,0 ng/ul
|
| I22-2 | 100,4 ng/ul
|
| I22-3 | 281,3 ng/ul
|
| I23-1 | 55,2 ng/ul
|
| I23-2 | 47,1 ng/ul
|
| I23-3 | 84,8 ng/ul
|
| I15-1 | 80,7 ng/ul
|
| Ent4C5 | 27,6 ng/ul
|
| <partinfo>BBa_B0034</partinfo> | 50,4 ng/ul
|
| linker (<partinfo>BBa_K105012</partinfo>) | 39,3 ng/ul
|
| plux (<partinfo>BBa_R0062</partinfo>) | 45,1 ng/ul
|
Digestion of:
| Culture | Kind | Final reaction volume (ul) | DNA (ul) | H20 (ul) | Enzyme 1 | Enzyme 2 | Buffer H
|
| I20-1 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-2 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-3 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-4 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-5 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-6 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-7 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-8 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-9 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-10 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-11 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I20-12 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-1 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-2 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-3 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-4 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-5 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-6 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-7 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-8 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-9 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-10 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-11 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-12 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-13 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-14 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-15 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-16 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I21-17 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I22-1 | Insert/Screening | 25 | 17 | 3,5 | 1 XbaI | 1 PstI | 2,5
|
| I22-2 | Insert/Screening | 25 | 20,5 | 0 | 1 XbaI | 1 PstI | 2,5
|
| I22-3 | Insert/Screening | 25 | 7,1 | 13,4 | 1 XbaI | 1 PstI | 2,5
|
| I23-1 | Insert/Screening | 25 | 10 | 10,5 | 1 XbaI | 1 PstI | 2,5
|
| I23-2 | Insert/Screening | 25 | 10 | 10,5 | 1 XbaI | 1 PstI | 2,5
|
| I23-3 | Insert/Screening | 25 | 10 | 10,5 | 1 XbaI | 1 PstI | 2,5
|
| I15-1 | insert | 25 | 20,5 | 0 | 1EcoRI | 1 PstI | 2,5
|
| Entero4C5 | Vector | 25 | 20,5 | 0 | 1 EcoRI | 1 PstI | 2,5
|
| <partinfo>BBa_B0034</partinfo> | Insert | 25 | 20,5 | 0 | 1 SpeI | 1 PstI | 2,5
|
| <partinfo>BBa_K105012</partinfo> | Insert | 25 | 20,5 | 0 | 1 SpeI | 1 PstI | 2,5
|
| <partinfo>BBa_R0062</partinfo> | Insert | 25 | 20,5 | 0 | 1 SpeI | 1 PstI | 2,5
|
| PhaP 10-S | Insert | 30+10° | 26 | 0 +7 | 1+1 XbaI | 1+1 SpeI | 3+1
|
| PhaP S-S | Insert | 30+10° | 26 | 0 +7 | 1+1 XbaI | 1+1 SpeI | 3+1
|
| <partinfo>pSB1A2</partinfo> | Insert | 25 | 14 | 6,5 | 1XbaI | 1 SpeI | 2,5
|
°(after '+' added after 2 hours and incubated for further 2 hours)
Digestions were incubated at 37°C for 3 hours. A big gel and 2 medium gel were prepared, samples were loaded and gel ran/cut.
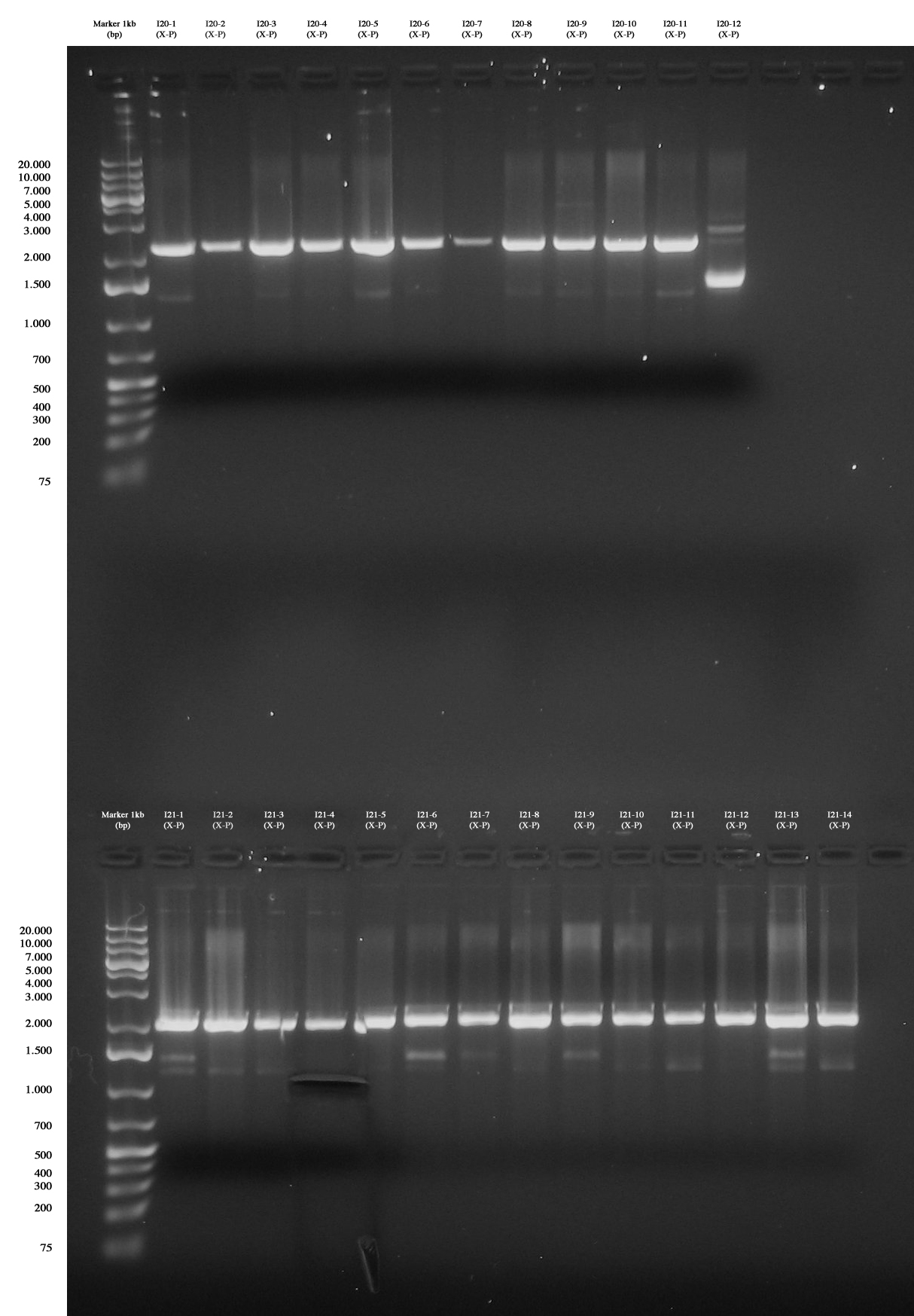 Up: Screening for I20-1..12 (PhaP 10-S (X-P)): all colonies are negative :( Down: Screening for I21-1..14 (PhaP S-S (X-P)): all colonies are negative, except for I21-4 gel run/cut and purified (we ran gel again after the cut). |
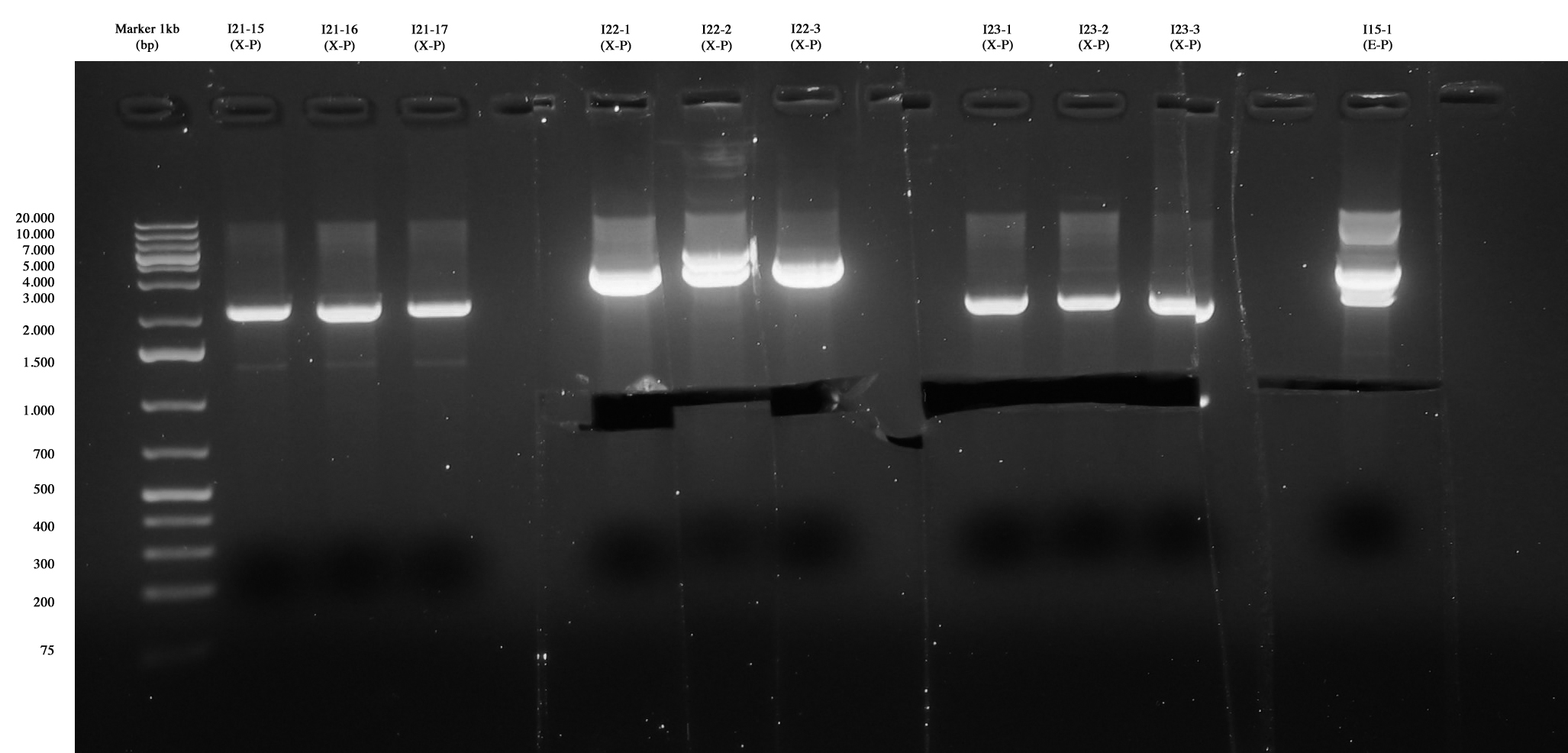 Screening for I21-15..17, I22, I23 and I15-1. The three colonies of I21 were negative, I22 and I23 were positive, so they were excided and purified. I15-1 was excided, even if extra-bands were observed. We use it for ligation because sequencing is ok! |
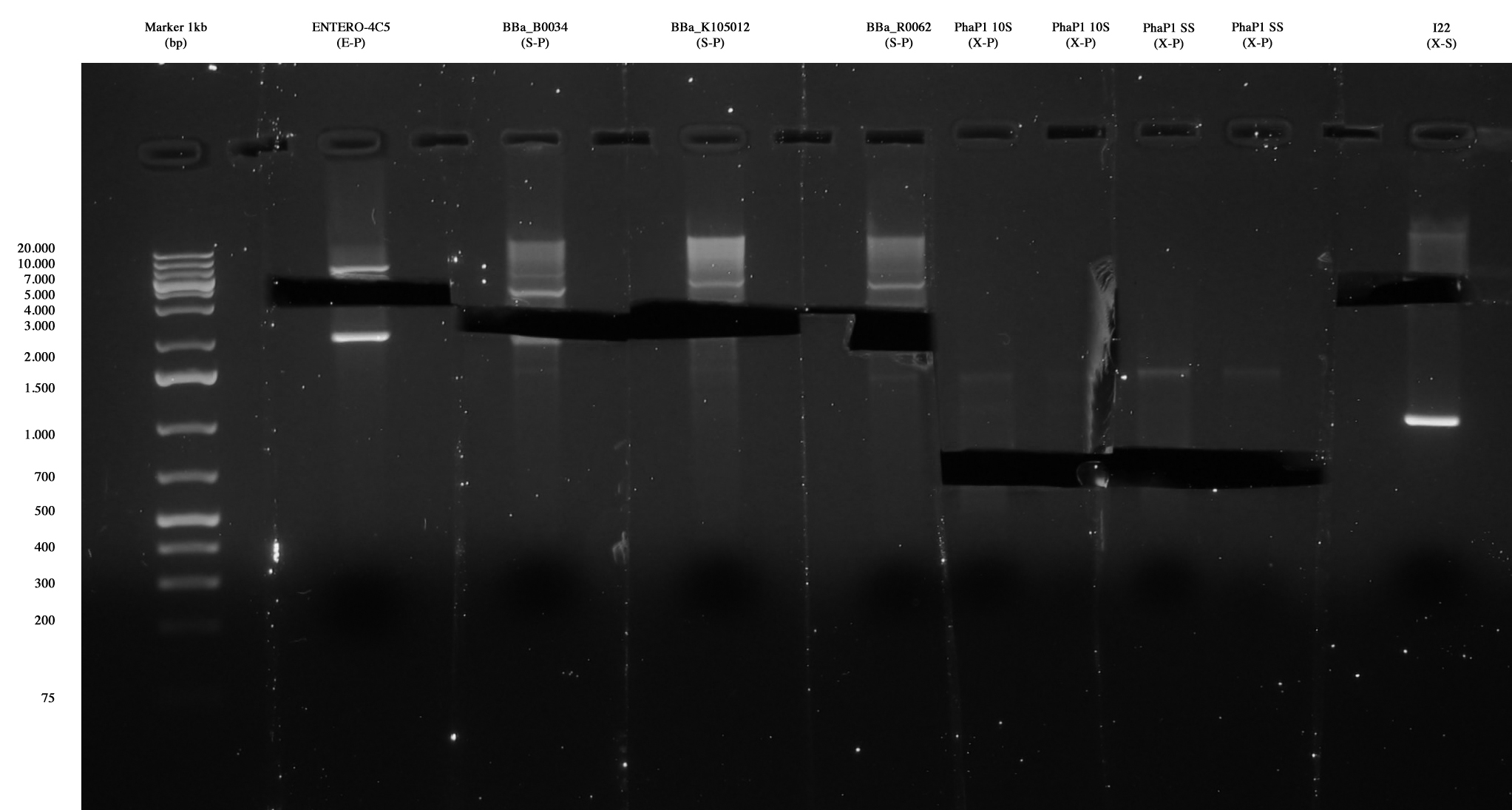 Ent4C5, <partinfo>BBa_B0034</partinfo>, <partinfo>BBa_K105012</partinfo> and <partinfo>BBa_R0062</partinfo> were extracted and purified. Also PhaP 10-S (X-S) and PhaP S-S (X-S) were extracted and purified, in order to repeat the ligation with a vector, since one only colony was positive for I21 and no colony was positive for I20. <partinfo>pSB1A2</partinfo> vector cut X-S was also gel extracted and purified. |
After purification, digested DNA was quantified as follows:
| I21-4 (X-P) | 1,7 ng/ul
|
| I22-1 (X-P) | 11,7 ng/ul
|
| I22-2 (X-P) | 6,1 ng/ul
|
| I22-3 (X-P) | 11,3 ng/ul
|
| I23-1 (X-P) | 4,5 ng/ul
|
| I23-2 (X-P) | 3,1 ng/ul
|
| I23-3 (X-P) | 5,6 ng/ul
|
| I15-1 (E-P) | 5,2 ng/ul
|
| <partinfo>pSB4C5</partinfo> (E-P) | 6,0 ng/ul
|
| <partinfo>BBa_B0034</partinfo> (E-P) | 20,9 ng/ul
|
linker (<partinfo>BBa_K105012</partinfo>)
(E-P) | 17,6 ng/ul
|
pLux (<partinfo>BBa_R0062</partinfo>)
(S-P) | 16,8 ng/ul
|
| PhaP 10-S (X-S) | 14,0 ng/ul
|
| PhaP S-S (X-S) | 13,0 ng/ul
|
| <partinfo>pSB1A2</partinfo> (X-S) | 18,6 ng/ul
|
Ligation of:
- I15_4C5= I15-1 (E-P)+pSB4C5(E-P)
- I24=I22-1(X-P)+Plux(S-P)
- I26=linker(S-P)+I21-4(X-P)
- I20_M=PhaP 10-S (X-S)+pSB1A2 (X-S) not dephosphorylized
- I21_M=PhaP S-S (X-S)+pSB1A2 (X-S) not dephosphorylized
- I20_A=PhaP 10-S (X-S)+pSB1A2 (X-S) dephosphorylized with antarctic phosphatase
- I21_A=PhaP S-S (X-S)+pSB1A2 (X-S) dephosphorylized with antarctic phosphatase
- I20_C=PhaP 10-S (X-S)+pSB1A2 (X-S) dephosphorylized with alkaline phosphatase (from calf intestine)
- I21_C=PhaP S-S (X-S)+pSB1A2 (X-S) dephosphorylized with alkaline phosphatase (from calf intestine)
Ligations were incubated ON at 16°C and tomorrow will be transformed.
Colonies transformed with RING, F2620-4C5, Nothing and plated on proper agar plates were still too little to check efficiency of MC123/008 and MG123/008 competent cells. So we let them grow another day and night at 30°C.
August, 11th
Plates grown at 30°C showed the following results:
| | MC123 | MC008 | MG123 | MG008
|
| F2620-4C5 | Grown (OK) | Grown (OK) | Grown (OK - but only ~10 colonies) | Grown (OK)
|
| RING | Not grown (OK) | Not plated | Not grown (OK) | Grown (OK)
|
| NOTHING | Not grown (OK) | Not grown (OK) | Grown (???) | Not grown (OK)
|
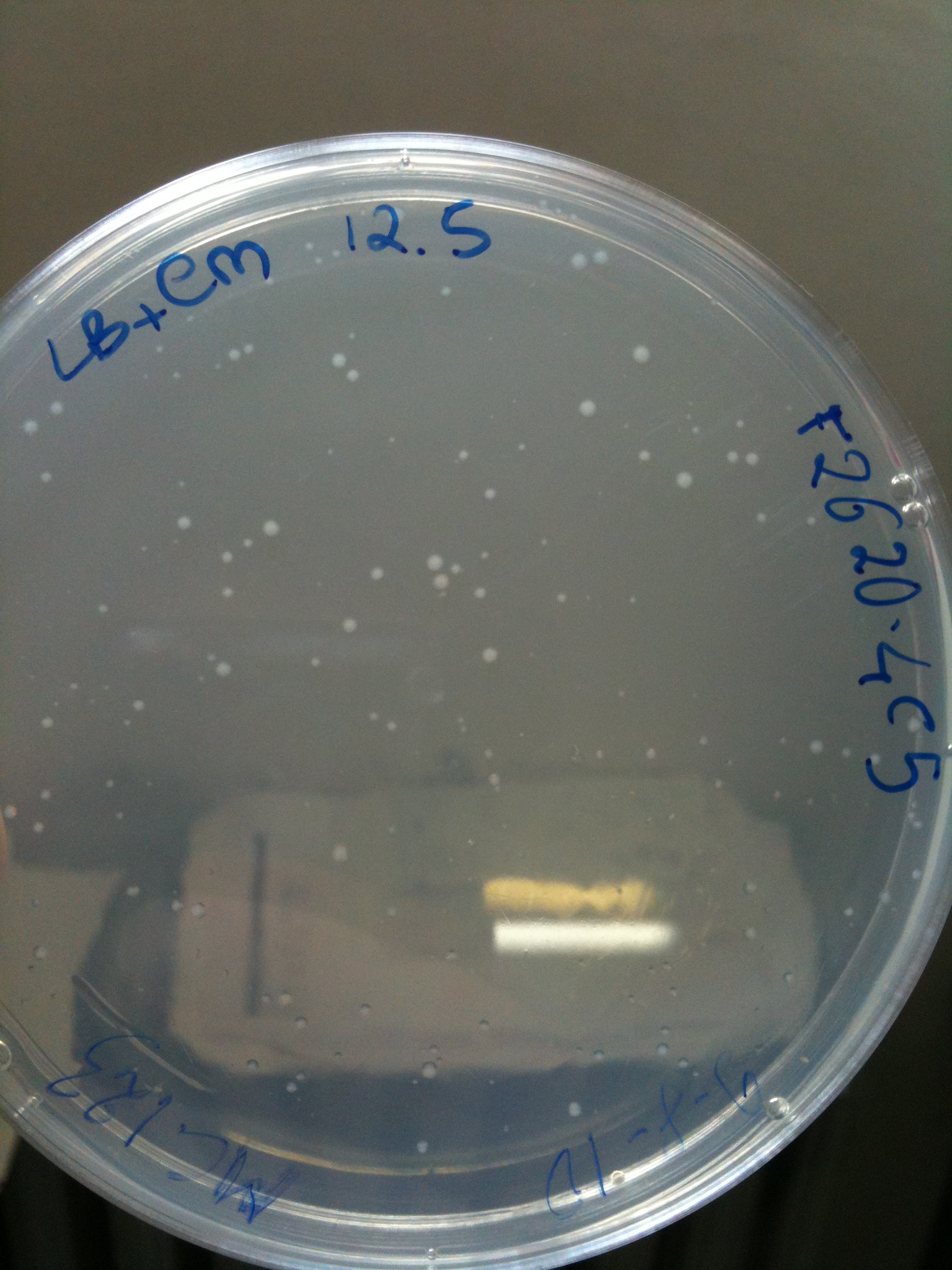 MC123 transformed with F2620-4C5 (OK) |  MC123 transformed with RING (OK) |  MC123 transformed with NOTHING (OK) |
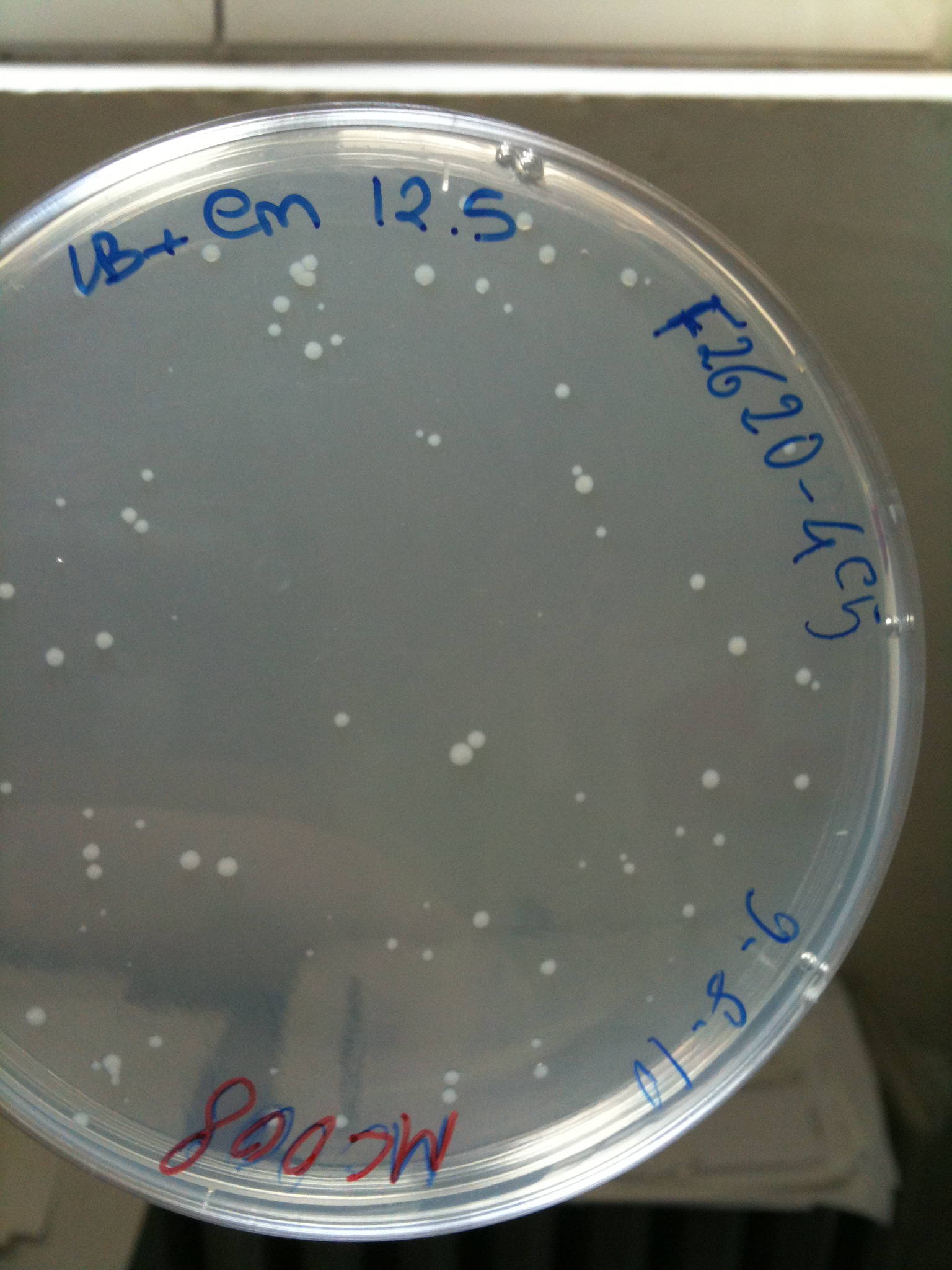 MC008 transformed with F2620-4C5 (OK) | MC008 transformed with RING
(Not plated) | 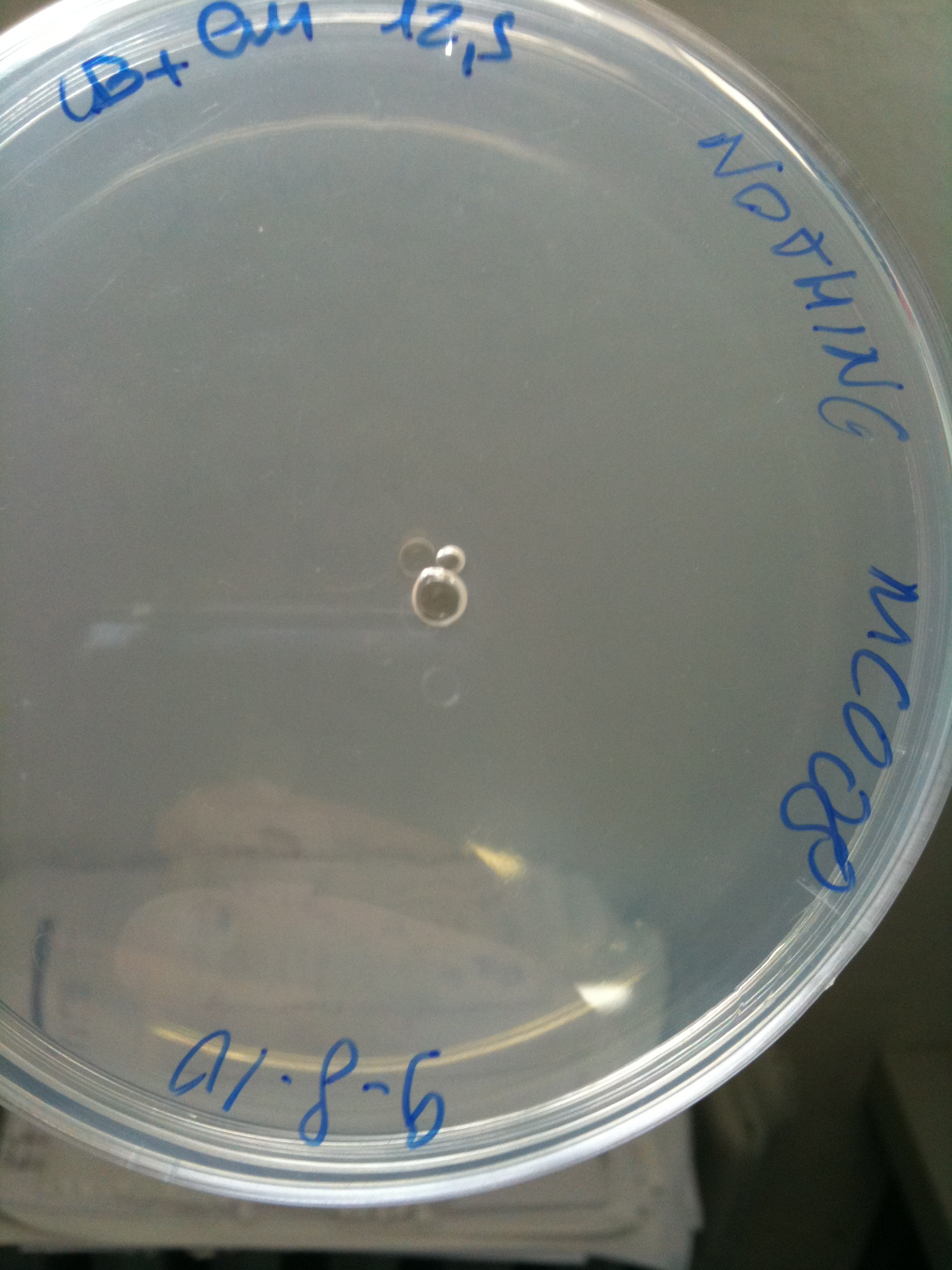 MC008 transformed with NOTHING (OK) |
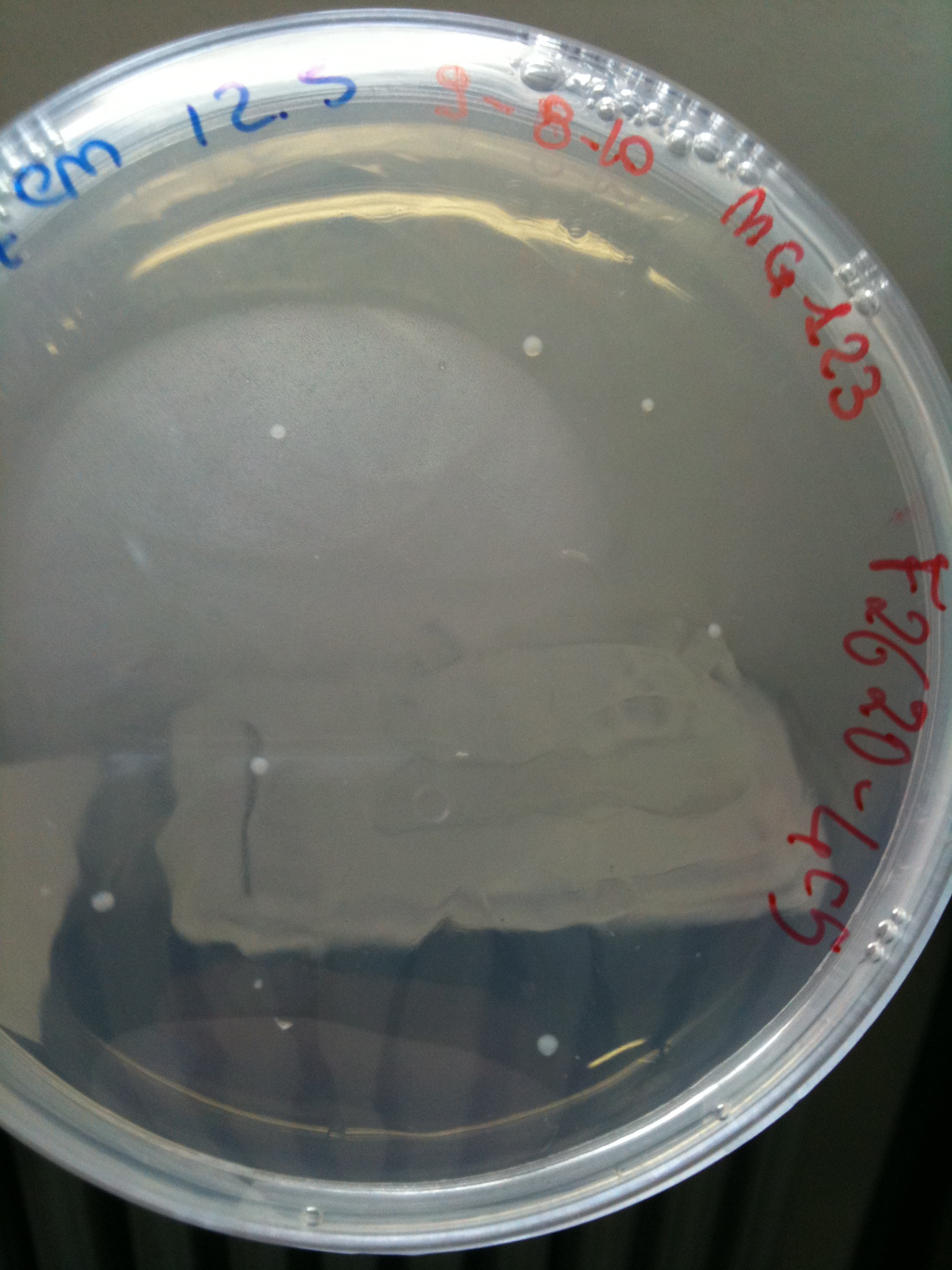 MG123 transformed with F2620-4C5 (OK - but only ~10 colonies) | 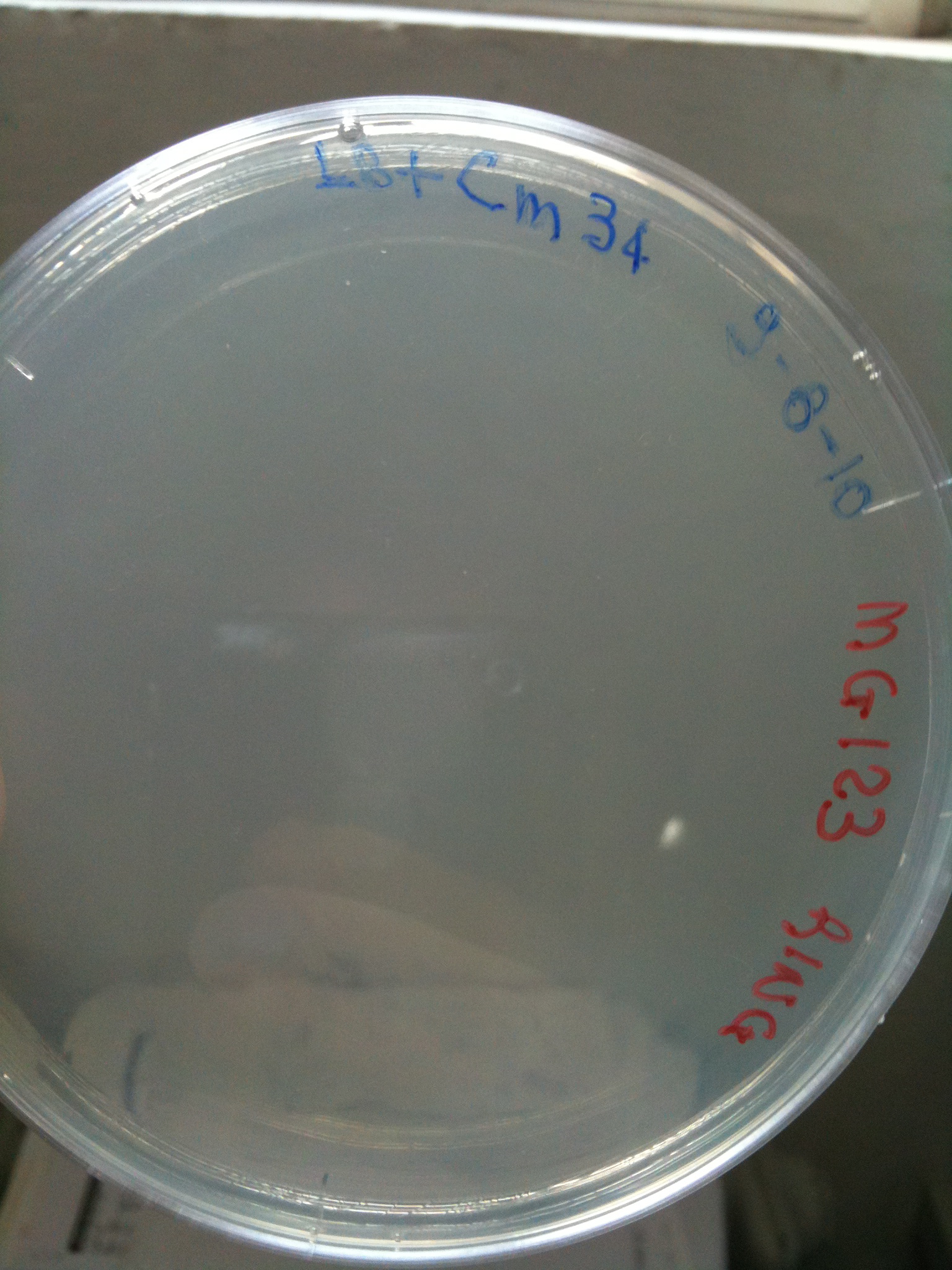 MG123 transformed with RING (OK) | 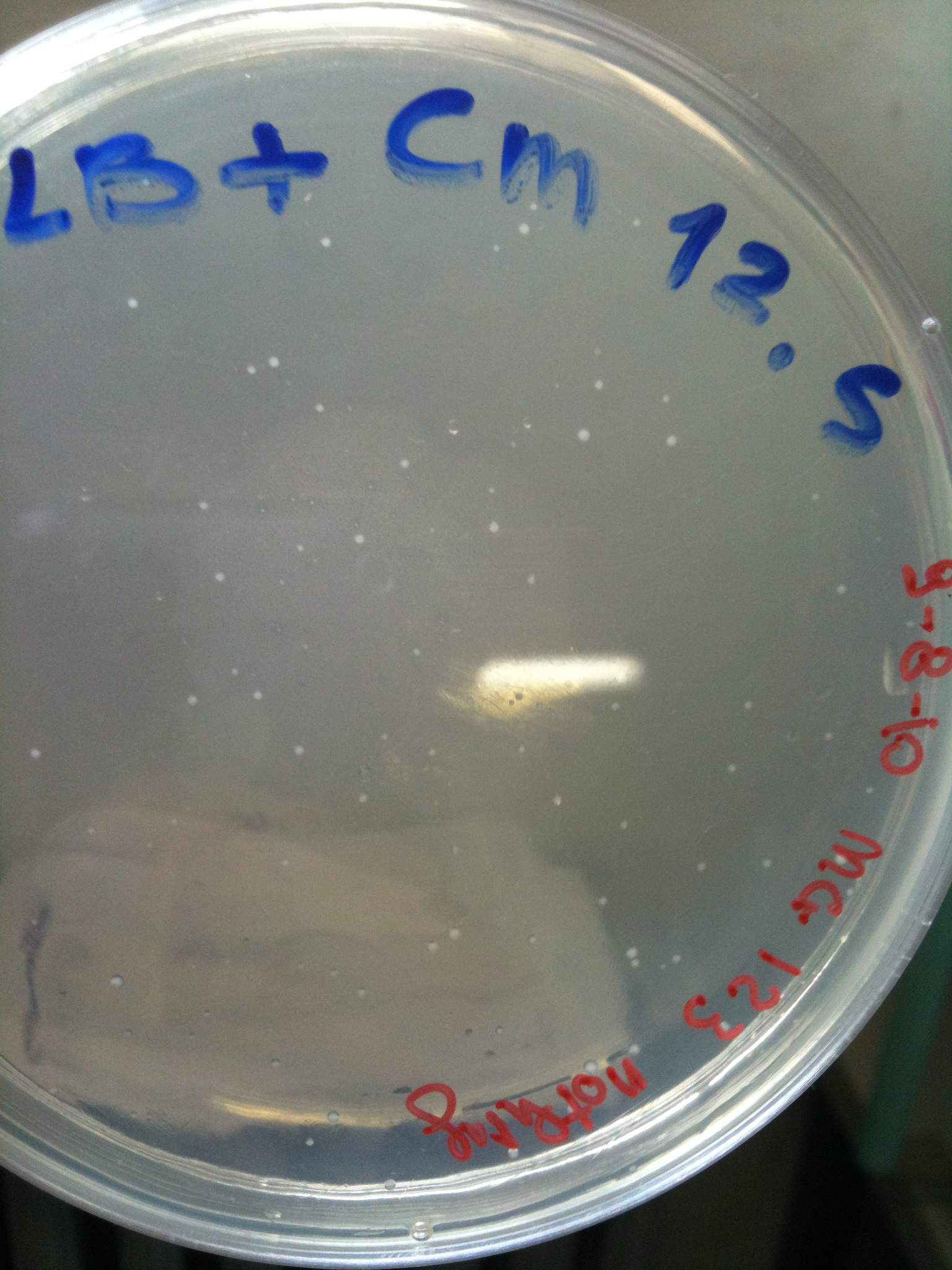 MG123 transformed with NOTHING (???) |
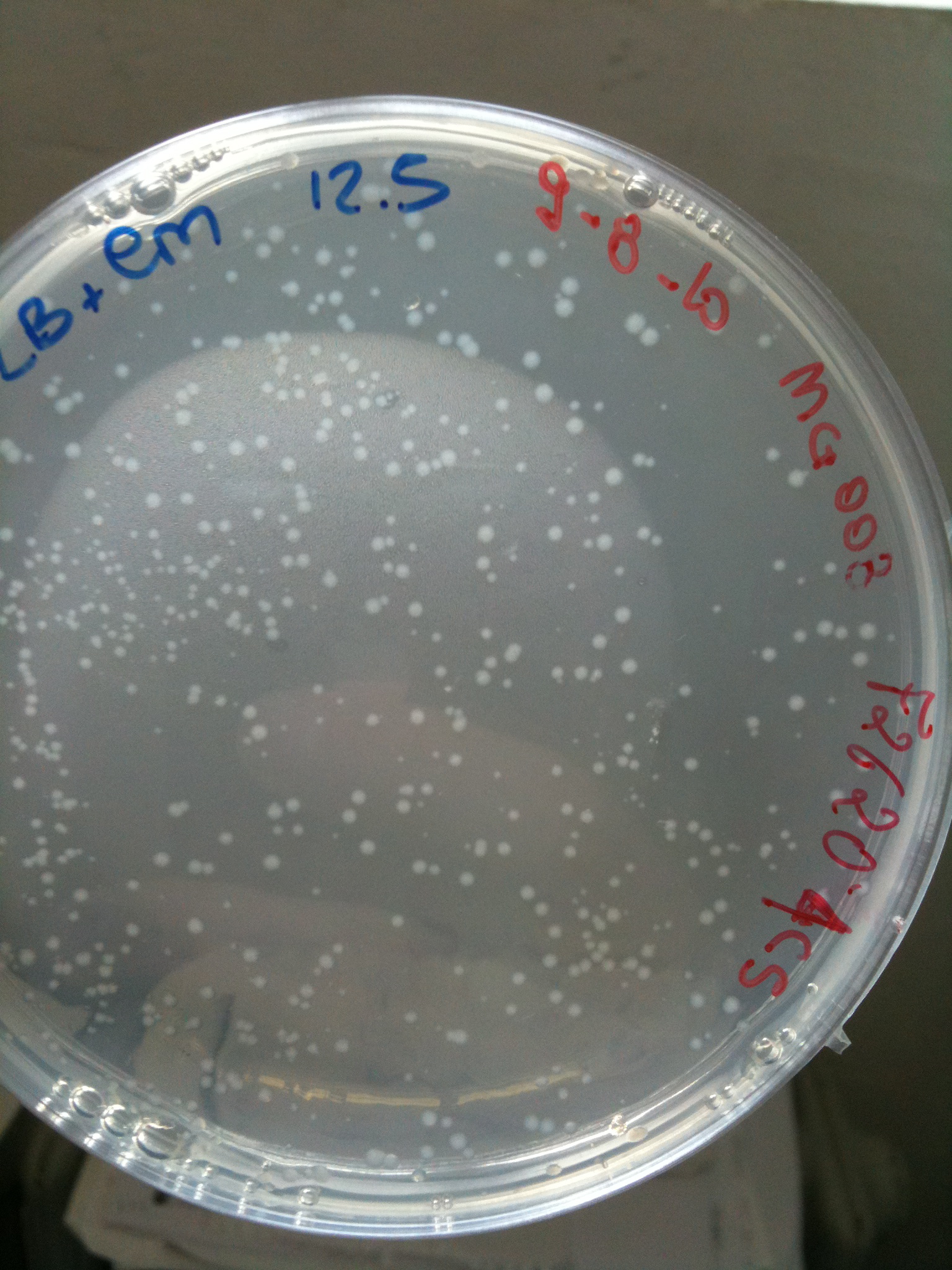 MG008 transformed with F2620-4C5 (OK) | 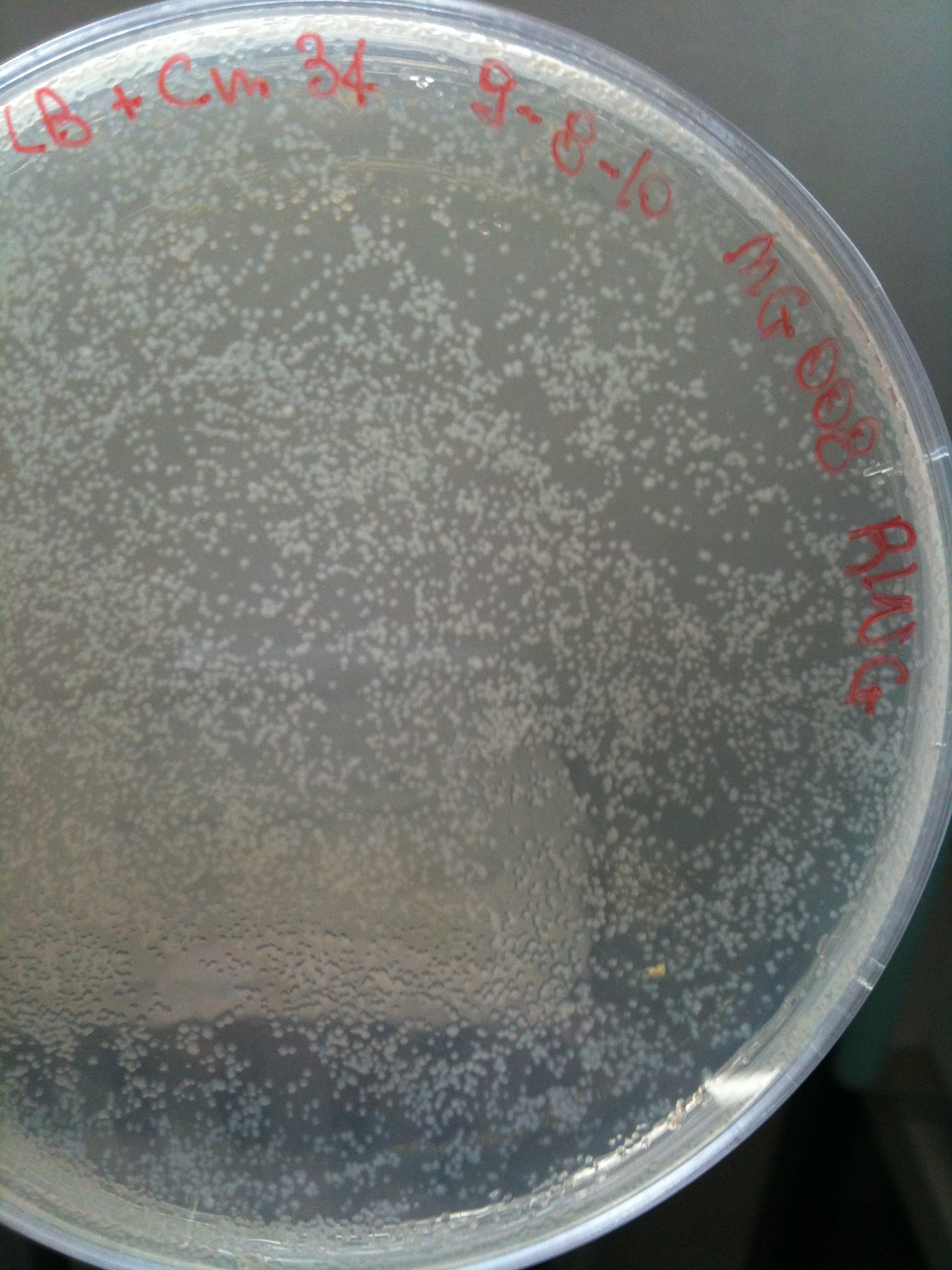 MG008 transformed with RING (OK) | 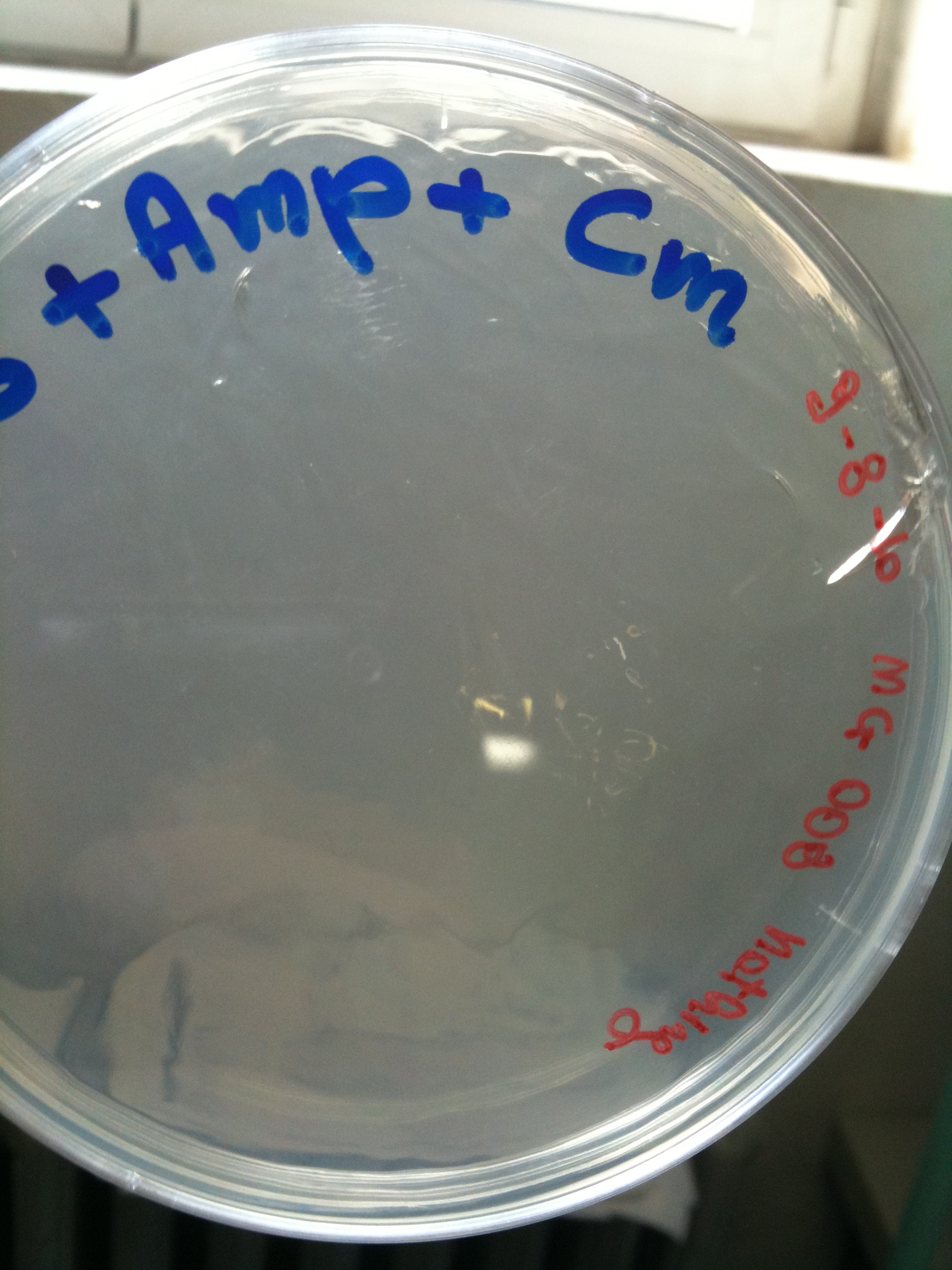 MG008 transformed with NOTHING (OK) |
So we decided to repeat transformation for
- MC008: F2620-4C5 (Cm 12,5 ug/ml), RING (Cm 34 ug/ml)
- MG123: F2620-4C5 (Cm 12,5 ug/ml), RING (Cm 34 ug/ml), NOTHING (12,5 ug/ml)
We transformed 1 ul (~4 ng of DNA) and plates were let grow at 30°C for about twenty hours.
August, 12th
August, 13th
August, 14th
August, 15th
|
|