Team:TU Delft/Modeling/HC regulation/Sensitivity
From 2010.igem.org
Sensitivity analysis
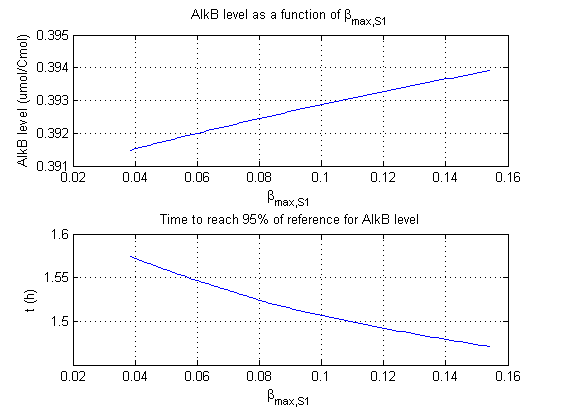
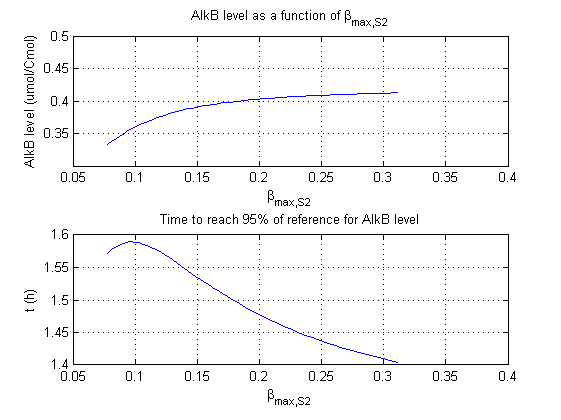
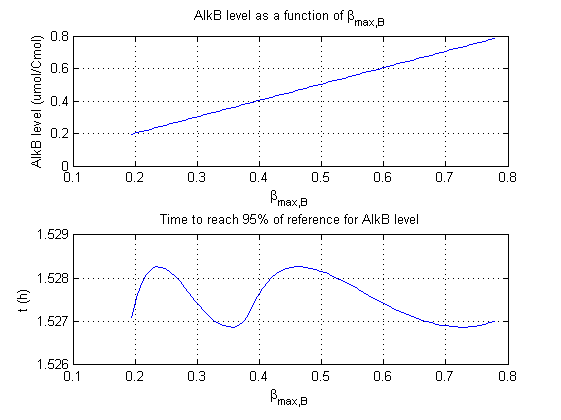
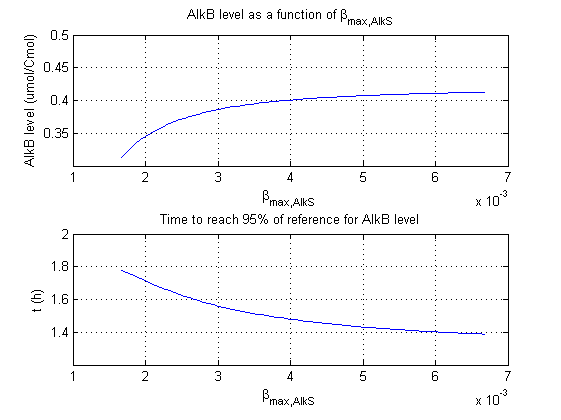
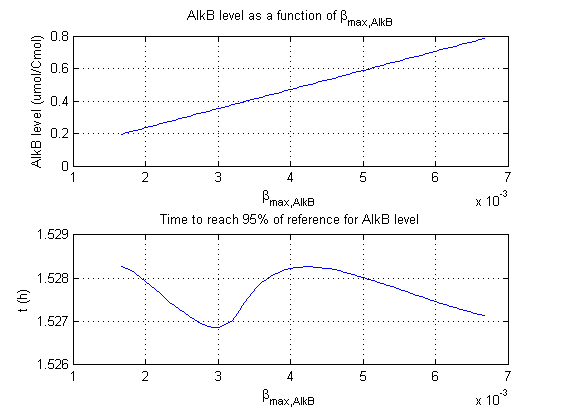
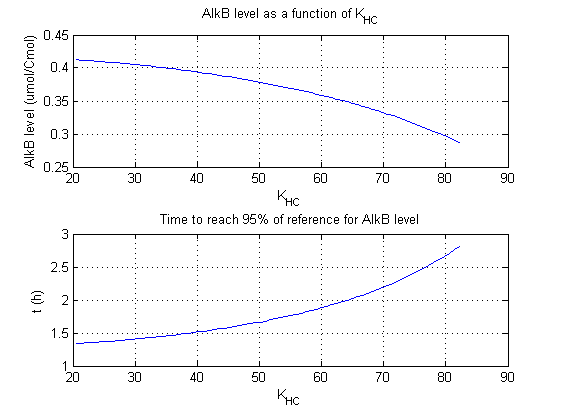
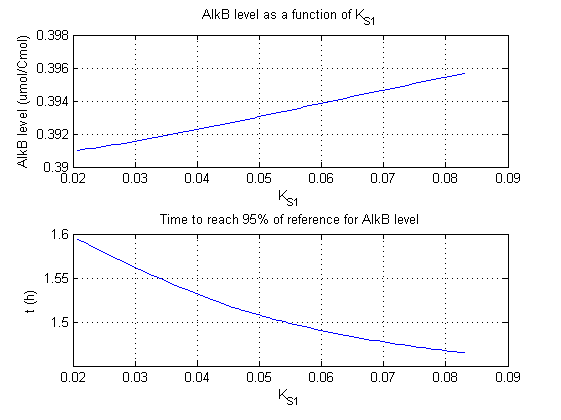
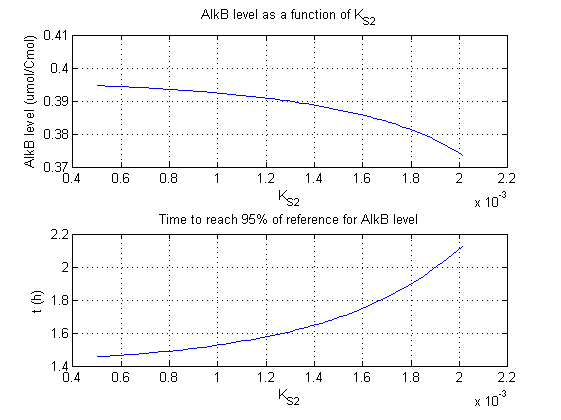
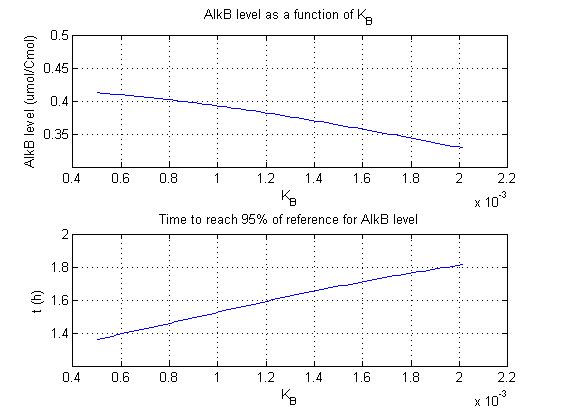
For all beta parameters (lumped variables for transcriptions speed, translations speed, promoter strength and rbs strength) and all K parameters (equilibrium constant for binding of AlkS with alkanes and coefficients for the activity of promoters) a sensitivity analysis has been performed. This was done by varying the parameters from 50% of their original values to 200% of their original values.
For this range of parameter values it is checked how much the steady state concentration of AlkB is and how long it takes for 95% of the steady state concentration of AlkB to be reached from steady state values for no alkanes to steady state values for 1 µM of alkanes.
In the figures below are the results for this sensitivity analysis.
These results are summarized in table 1. A '0' means insensitive, a '+' means somewhat sensitive and a '++' is very sensitive.
Table 1; results for sensitivity analysis for the gene regulation model
| Parameter | Sensitive for steady state concentration | Sensitive for reaching steady state concentration |
| βmaxS1 | 0 | + |
| βmaxS2 | + | + |
| βmaxB | ++ | 0 |
| βmaxAlkS | + | + |
| βmaxAlkB | ++ | 0 |
| KHC | + | + |
| KS1 | 0 | + |
| KS2 | + | + |
| KB | + | + |
Go back to the model explanation here
Go back to the main modeling page here
 "
"