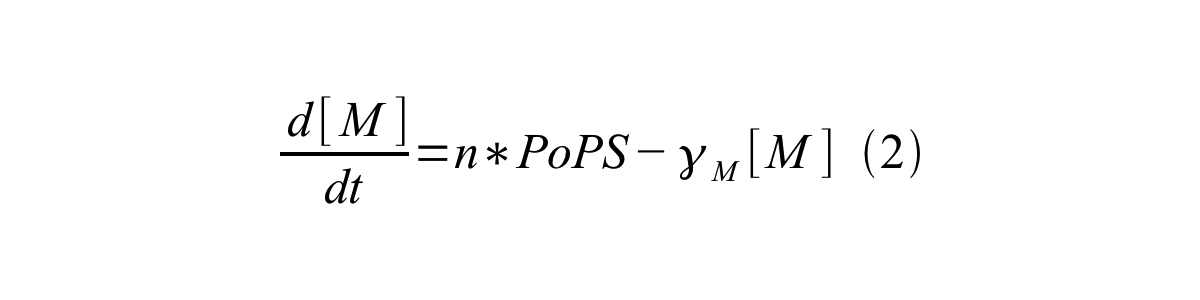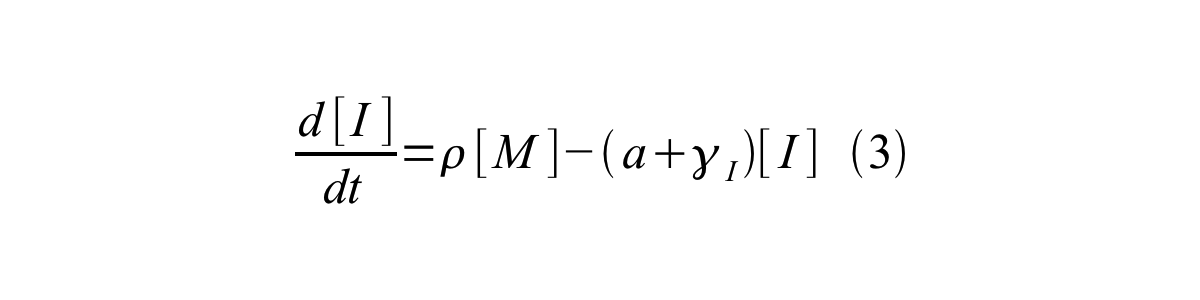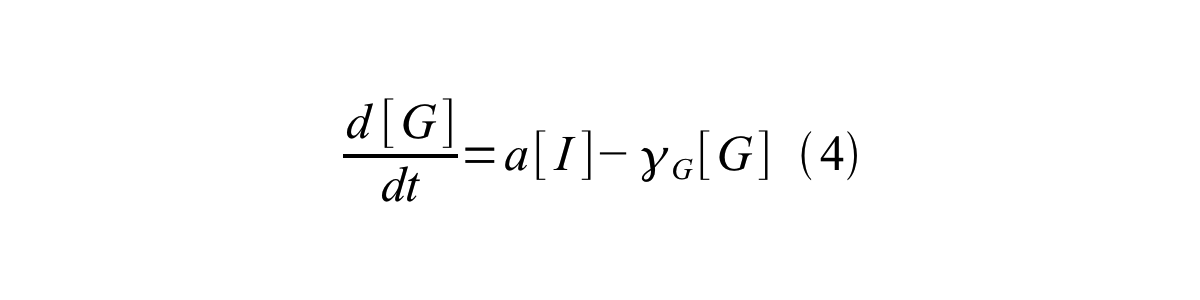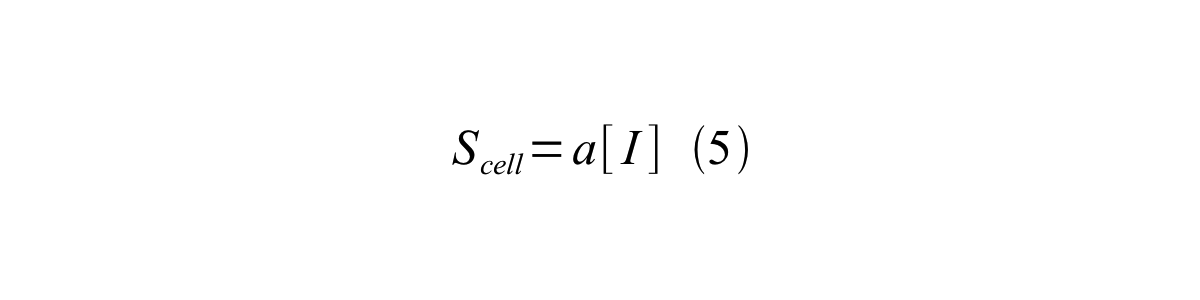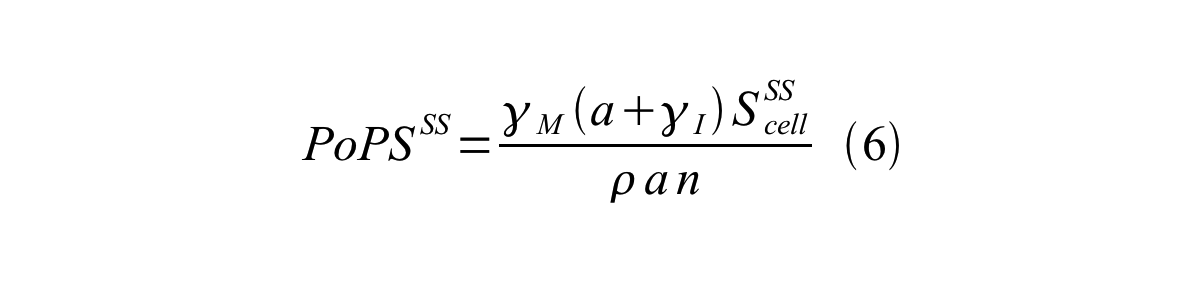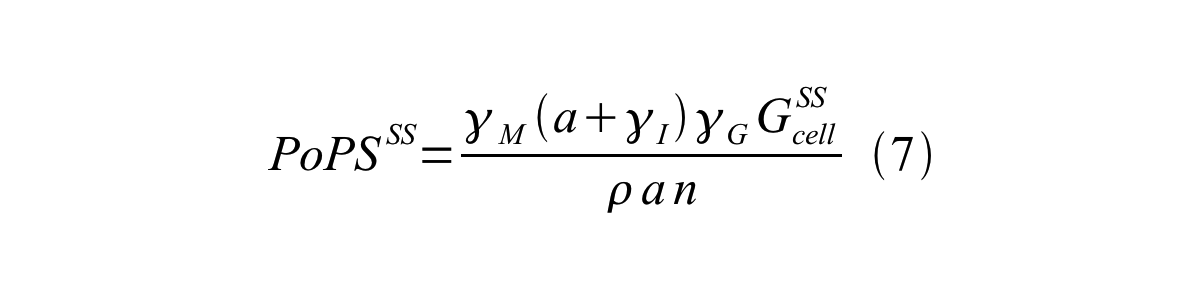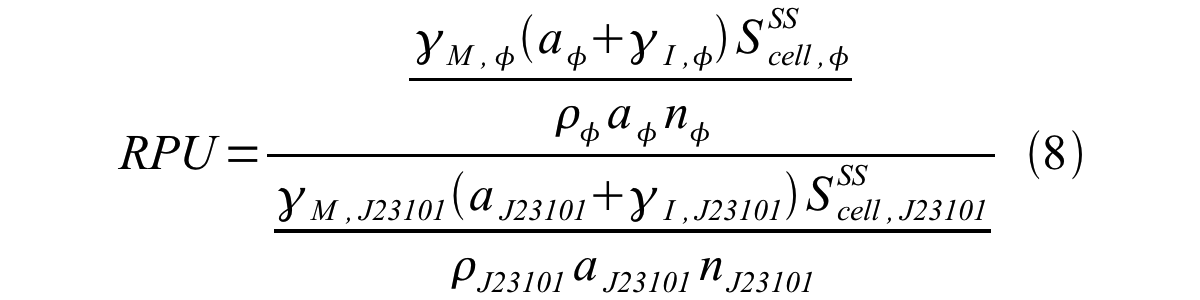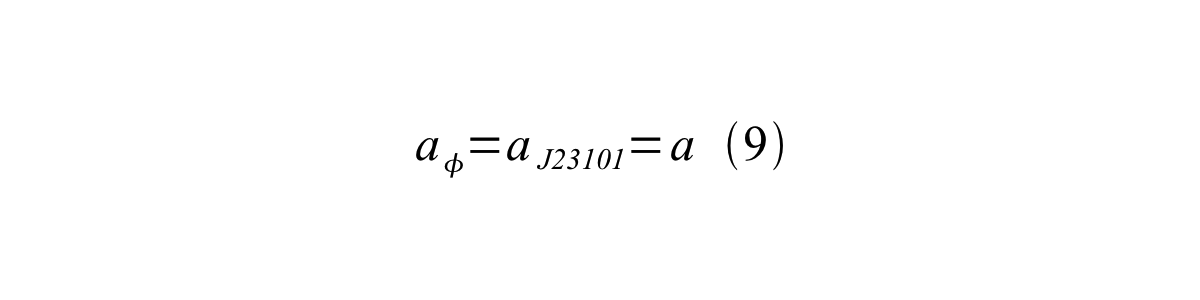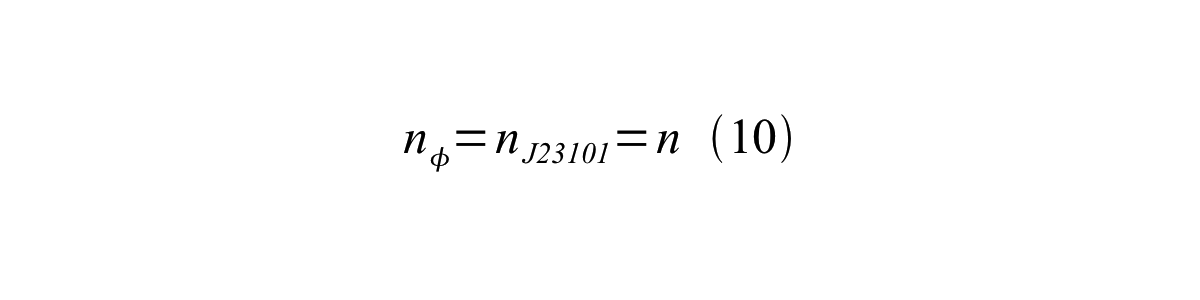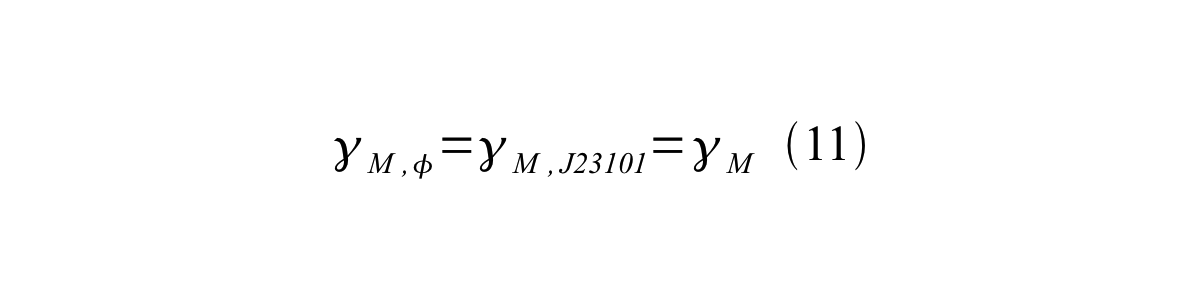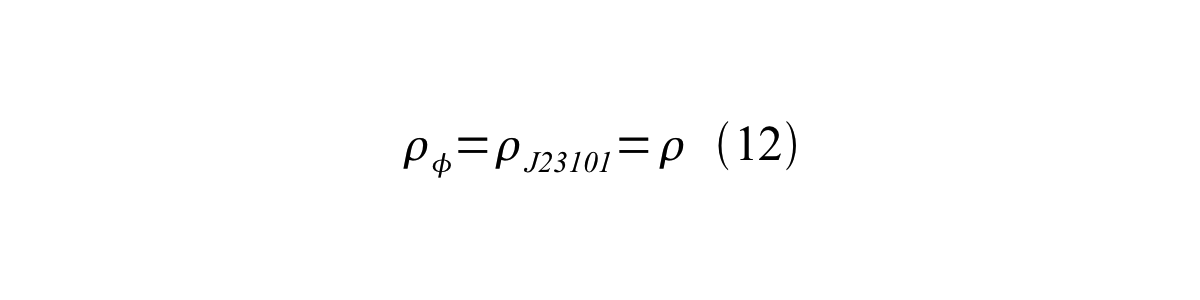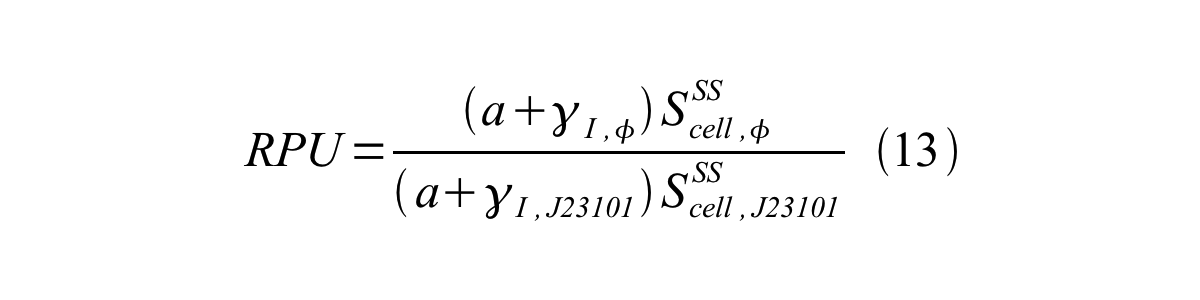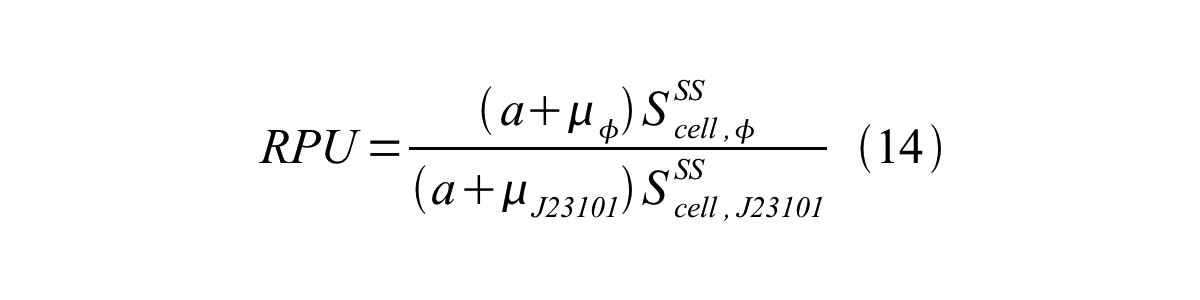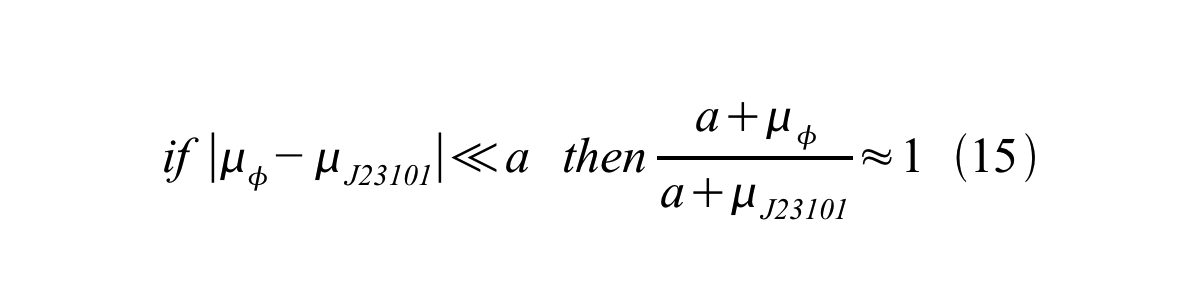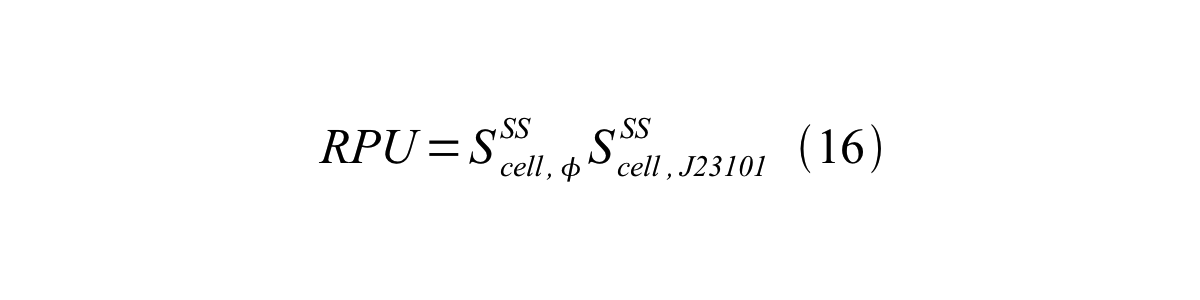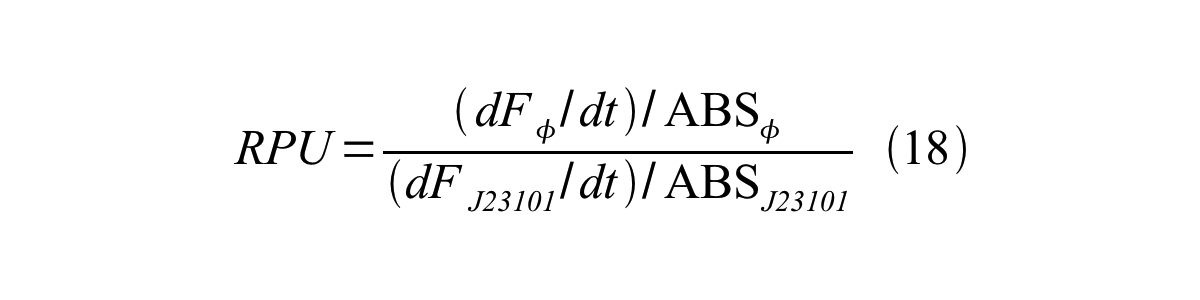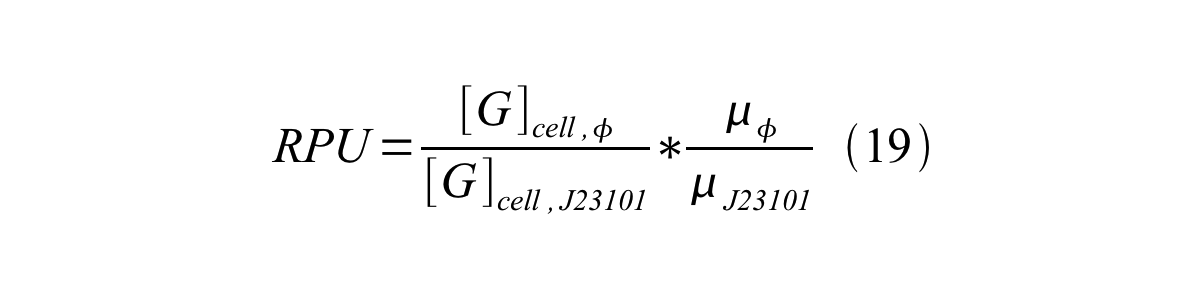Team:Kyoto/LearnMore
From 2010.igem.org
Contents |
LearnMore
R0011
In our project, we utilized BioBrick part of BBa_R0011. This is one of the inducible promoter parts in iGEM, which is often used by a lot of iGEM teams. R0011 is composed of a unique Lac promoter (Plac) whose promoter activity is stronger than an ordinary Plac of lambda phage. First, we explain the ordinary Plac and subsequently the uniqueness of R0011.
Lac promoter of lambda phage
lacP of lambda phage is composed of the cI binding sites and Lac operator (LacO) which is bound by Lac inhibitor (LacI) (Figure 1). This promoter is controlled by the interaction of LacI and lactose as inducer. lacP is classified as a negatively regulated promoter. This means that lacP has the strong promoter activity in the absence of LacI because RNA polymerase can approach the promoter and constitutively transcribe the downstream genes (Figure 2.a). However, lacP decreases the activity with increasing LacI. At this time, LacI binds to the promoter and therefore RNA polymerase is prevented approaching the promoter. As a result, the downstream genes are hardly transcribed (Figure 2.b). When lactose is introduced, it binds to LacI and prevents LacI from binding to the promoter. Then RNA polymerase can approach the promoter and transcribes the downstream genes (Figure 2.c).
The uniqueness of R0011
As stated above, lacP of lambda phage has the cI binding sites and LacO (Figure 1). R0011 replaces cI binding sites with LacO. In other words, this part has two LacO, the original LacO and the extra LacO instead of cI binding sites (Figure 3). Nevertheless, this hybrid design has the strong promoter activity. Moreover, this part is repressed by LacI and induced by lactose as same as lacP of lambda phage.
Lysis Cassette
Sam7 Mutation
RPU (Relative Promoter Unit)
Overview
Though BioBrick parts are standardized in terms of how individual parts are physically assembled into multi-component systems, many of them are not standardized in the point of characterization.
Unfortunately, it is known that the absolute activity of BioBrick promoters varies across experimental conditions and measurement instruments [1]. However, Kelly et al. found that this variation could be reduced by measuring the activity of promoters relative to <partinfo>J23101</partinfo> which is a promoter as an in vivo reference standard for promoter activity by -50%. They defined a Relative Promoter Units (RPU) in order to report promoter characterization data in compatible units. In order to measure RPUs of promoters, the synthesis rate of Green Fluorescent Protein (GFP) encoded by mRNA transcribed from each promoter is observed.
RPU is defined by equation 1:
Derivation
PoPS (Polymerase Per Second) is the unit of absolute “promoter activity”. It is defined as the number of RNA polymerase molecules that pass by the final base pair of the promoter and continue along DNA as an elongation complex. PoPSSS is PoPS at the steady state of following system (d[M]/dt=0, d[I]/dt=0, d[G]/dt=0:
Where:
- [M] is the concentration of mRNA,
- [I] is the concentration of immature GFP,
- [G] is the concentration of mature GFP,
- γM is the mRNA degradation rate,
- a is the GFP maturation rate,
- γI is the degradation rate of immature GFP
- n is the copy number of the plasmid containing the promoter
- ρ is the rate of synthesis of immature GFP in absolute units of protein per second per mRNA.
By combining equation 2, 3, 4 and 5 at the steady state, equation 6 and 7 are obtained:
Here Scell is define as the per cell mature GFP synthesis term.
Here Gcell is define as the per cell mature GFP synthesis term.
From equation 1 and 6 RPU is described as equation8.
If the test promoter φ and the reference standard promoter are measured under the same culture conditions and both promoters are carried on the same backbone plasmid, following equations are approved.
From equation 8, 9, 10, 11, and 12, equation 13 is accomplished.
In addition, immature GFP is stable so that protein degradation is negligible compared to dilution due to cell growth (γI, φ=μφ and γI,,J23101=μJ23101, where μis the cellular growth rate). Therefore, equation 13 can be modified to equation 14:
And if the growth rates of both strains are almost same, equation 15 is approved.
Accordingly, we can make equation 16 from equation15.
ScellSS can be described by equation17:
Therefore, RPU is described by equation 18:
We can also establish equation20 from equation1, 6, 9, 10, 11 and 12.
RPU can be calculated from equation 18 or equation 19. Therefore, we have to observe GFP synthesis rate and ABS (OD600) of both strains or to measure GFP concentration per cell and growth rate of both strains to measure RPU.
Note
To see how to measure RPU practically, go Protocols
Reference
- [http://www.ncbi.nlm.nih.gov/pubmed/19298678 PMID: 19298678] Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D., Measuring the activity of BioBrick promoters using an in vivo reference standard., J Biol Eng. 2009 Mar 20;3:4.
 "
"